3FPU
 
 | |
1JTG
 
 | | CRYSTAL STRUCTURE OF TEM-1 BETA-LACTAMASE / BETA-LACTAMASE INHIBITOR PROTEIN COMPLEX | | 分子名称: | BETA-LACTAMASE INHIBITORY PROTEIN, BETA-LACTAMASE TEM, CALCIUM ION | | 著者 | Strynadka, N.C.J, Jensen, S.E, Alzari, P.M, James, M.N. | | 登録日 | 2001-08-20 | | 公開日 | 2001-10-17 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Crystal structure and kinetic analysis of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase.
Nat.Struct.Biol., 8, 2001
|
|
2KR6
 
 | |
6MSU
 
 | | Integrin alphaVBeta3 in complex with EETI-II 2.5F | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | van Agthoven, J.F, Arnaout, M.A. | | 登録日 | 2018-10-18 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Structural Basis of the Differential Binding of Engineered Knottins to Integrins alpha V beta 3 and alpha 5 beta 1.
Structure, 27, 2019
|
|
2KWF
 
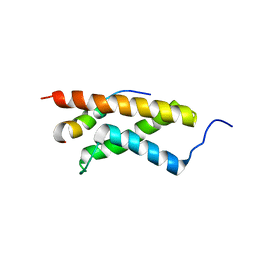 | | The structure of E-protein activation domain 1 bound to the KIX domain of CBP/p300 elucidates leukemia induction by E2A-PBX1 | | 分子名称: | CREB-binding protein, Transcription factor 4 | | 著者 | Denis, C.M, Chitayat, S, Plevin, M.J, Liu, S, Spencer, H.L, Ikura, M, LeBrun, D.P, Smith, S.P. | | 登録日 | 2010-04-08 | | 公開日 | 2011-07-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of E-protein activation domain 1 bound to the KIX domain of CBP/p300 elucidates leukemia induction by E2A-PBX1
To be Published
|
|
4EUZ
 
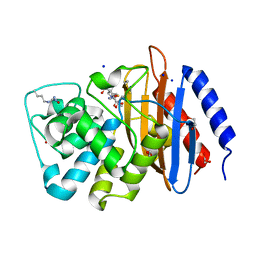 | | Crystal structure of serratia fonticola carbapenemase SFC-1 S70A-Meropenem complex | | 分子名称: | (4R,5S,6S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-6-[(1R)-1-hydroxyethyl]-4-methyl-7-oxo-1-azabicyclo[3.2.0]hept-2-ene-2-carboxylic acid, 1,2-ETHANEDIOL, Carbapenem-hydrolizing beta-lactamase SFC-1, ... | | 著者 | Fonseca, F, Spencer, J. | | 登録日 | 2012-04-25 | | 公開日 | 2012-05-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | The basis for carbapenem hydrolysis by class A beta-lactamases: a combined investigation using crystallography and simulations.
J.Am.Chem.Soc., 134, 2012
|
|
3BT4
 
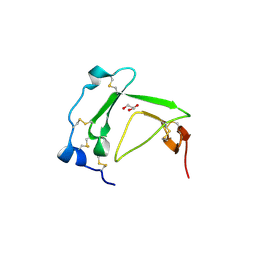 | | Crystal Structure Analysis of AmFPI-1, fungal protease inhibitor from Antheraea mylitta | | 分子名称: | Fungal protease inhibitor-1, GLYCEROL | | 著者 | Roy, S, Aravind, P, Madhurantakam, C, Ghosh, A.K, Sankarananarayanan, R, Das, A.K. | | 登録日 | 2007-12-27 | | 公開日 | 2008-12-30 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of a fungal protease inhibitor from Antheraea mylitta
J.Struct.Biol., 166, 2009
|
|
1GFI
 
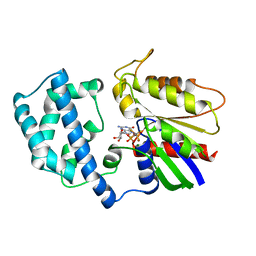 | | STRUCTURES OF ACTIVE CONFORMATIONS OF GI ALPHA 1 AND THE MECHANISM OF GTP HYDROLYSIS | | 分子名称: | GUANINE NUCLEOTIDE-BINDING PROTEIN G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Coleman, D.E, Berghuis, A.M, Sprang, S.R. | | 登録日 | 1994-11-11 | | 公開日 | 1995-03-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis.
Science, 265, 1994
|
|
7DH0
 
 | | Activity optimized complex I (open form) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | 登録日 | 2020-11-12 | | 公開日 | 2022-05-18 | | 最終更新日 | 2025-09-17 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DGS
 
 | | Activity optimized supercomplex state3 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | 登録日 | 2020-11-12 | | 公開日 | 2022-05-18 | | 最終更新日 | 2025-09-17 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DGQ
 
 | | Activity optimized supercomplex state1 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | 登録日 | 2020-11-12 | | 公開日 | 2022-05-18 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DGR
 
 | | Activity optimized supercomplex state2 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | 登録日 | 2020-11-12 | | 公開日 | 2022-05-18 | | 最終更新日 | 2025-09-17 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DKF
 
 | | Activity optimized supercomplex state4 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | 登録日 | 2020-11-24 | | 公開日 | 2022-05-18 | | 最終更新日 | 2025-09-17 | | 実験手法 | ELECTRON MICROSCOPY (8.3 Å) | | 主引用文献 | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
4L7D
 
 | | Structure of keap1 kelch domain with (1S,2R)-2-{[(1S)-5-methyl-1-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid | | 分子名称: | (1S,2R)-2-{[(1S)-5-methyl-1-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1 | | 著者 | Jnoff, E, Brookfield, F, Albrecht, C, Barker, J.J, Barker, O, Beaumont, E, Bromidge, S, Brooks, M, Ceska, T, Courade, J.P, Crabbe, T, Duclos, S, Fryatt, T, Jigorel, E, Kwong, J, Sands, Z, Smith, M.A. | | 登録日 | 2013-06-13 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Binding Mode and Structure-Activity Relationships around Direct Inhibitors of the Nrf2-Keap1 Complex.
Chemmedchem, 9, 2014
|
|
7DGZ
 
 | | Activity optimized complex I (closed form) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | 登録日 | 2020-11-12 | | 公開日 | 2022-05-18 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
1F0E
 
 | | Cecropin A(1-8)-magainin 2(1-12) modified gig to P in dodecylphosphocholine micelles | | 分子名称: | CECROPIN A-MAGAININ 2 HYBRID PEPTIDE | | 著者 | Oh, D, Shin, S.Y, Lee, S, Kim, Y. | | 登録日 | 2000-05-16 | | 公開日 | 2000-06-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Role of the hinge region and the tryptophan residue in the synthetic antimicrobial peptides, cecropin A(1-8)-magainin 2(1-12) and its analogues, on their antibiotic activities and structures.
Biochemistry, 39, 2000
|
|
3WWX
 
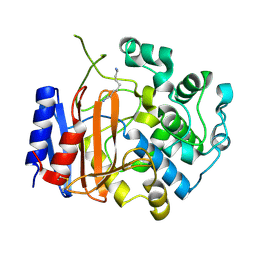 | | Crystal structure of D-stereospecific amidohydrolase from Streptomyces sp. 82F2 | | 分子名称: | OCTANE 1,8-DIAMINE, S12 family peptidase | | 著者 | Arima, J, Nagano, S, Hino, T, Shimone, K, Isoda, Y, Mori, N. | | 登録日 | 2014-07-03 | | 公開日 | 2015-07-08 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Crystal structure of D-stereospecific amidohydrolase from Streptomyces sp. 82F2 - insight into the structural factors for substrate specificity.
Febs J., 283, 2016
|
|
3ET1
 
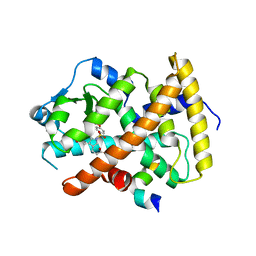 | |
3C33
 
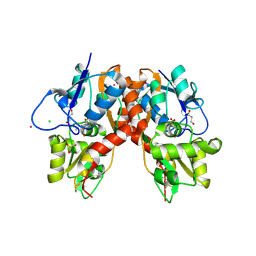 | |
4HLY
 
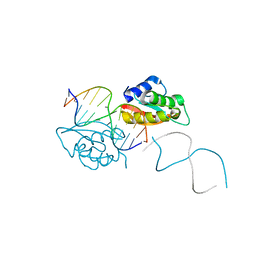 | |
2X7U
 
 | | Structures of human carbonic anhydrase II inhibitor complexes reveal a second binding site for steroidal and non-steroidal inhibitors. | | 分子名称: | (9BETA,14BETA,17BETA)-17-HYDROXY-2-METHOXYESTRA-1,3,5(10)-TRIEN-3-YL SULFAMATE, CARBONIC ANHYDRASE 2, GLYCEROL, ... | | 著者 | Cozier, G.E, Leese, M.P, Lloyd, M.D, Baker, M.D, Thiyagarajan, N, Acharya, K.R, Potter, B.V.L. | | 登録日 | 2010-03-03 | | 公開日 | 2010-03-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structures of Human Carbonic Anhydrase II/Inhibitor Complexes Reveal a Second Binding Site for Steroidal and Non-Steroidal Inhibitors.
Biochemistry, 49, 2010
|
|
2P73
 
 | | crystal structure of a glycosyltransferase involved in the glycosylation of the major capsid of PBCV-1 | | 分子名称: | MANGANESE (II) ION, Putative glycosyltransferase (Mannosyltransferase) involved in glycosylating the PBCV-1 major capsid protein, URIDINE-5'-DIPHOSPHATE | | 著者 | Zhang, Y, Xiang, Y, Van Etten, J.L, Rossmann, M.G. | | 登録日 | 2007-03-19 | | 公開日 | 2007-08-21 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and function of a chlorella virus-encoded glycosyltransferase.
Structure, 15, 2007
|
|
1FXJ
 
 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE | | 著者 | Brown, K, Pompeo, F, Dixon, S, Mengin-Lecreulx, D, Cambillau, C, Bourne, Y. | | 登録日 | 2000-09-26 | | 公開日 | 2000-10-18 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily.
EMBO J., 18, 1999
|
|
4CIW
 
 | | Crystal structure of Mycobacterium tuberculosis type 2 dehydroquinase in complex with (1R,4R,5R)-1,4,5-trihydroxy-3-(2-hydroxy)ethylcyclohex-2-ene-1-carboxylic acid | | 分子名称: | (1R,4R,5R)-1,4,5-trihydroxy-3-(2-hydroxy)ethylcyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, SODIUM ION, ... | | 著者 | Otero, J.M, Llamas-Saiz, A.L, Lamb, H, Hawkins, A.R, Blanco, B, Sedes, A, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | 登録日 | 2013-12-17 | | 公開日 | 2014-04-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Exploring the water-binding pocket of the type II dehydroquinase enzyme in the structure-based design of inhibitors.
J. Med. Chem., 57, 2014
|
|
2P72
 
 | | crystal structure of a glycosyltransferase involved in the glycosylation of the major capsid of PBCV-1 | | 分子名称: | MANGANESE (II) ION, Putative glycosyltransferase (Mannosyltransferase) involved in glycosylating the PBCV-1 major capsid protein, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | 著者 | Zhang, Y, Xiang, Y, Van Etten, J.L, Rossmann, M.G. | | 登録日 | 2007-03-19 | | 公開日 | 2007-08-21 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and function of a chlorella virus-encoded glycosyltransferase.
Structure, 15, 2007
|
|
