3NOA
 
 | |
7QSD
 
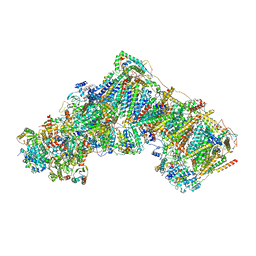 | | Bovine complex I in the active state at 3.1 A | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Bridges, H.R, Blaza, J.N, Yin, Z, Chung, I, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-03-08 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides.
Science, 379, 2023
|
|
4YOS
 
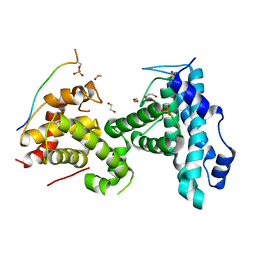 | | p107 pocket domain complexed with LIN52 peptide | | 分子名称: | 1,2-ETHANEDIOL, Protein lin-52 homolog, Retinoblastoma-like protein 1,Retinoblastoma-like protein 1, ... | | 著者 | Guiley, K.Z, Liban, T.J, Felthousen, J.G, Ramanan, P, Tripathi, S, Litovchick, L, Rubin, S.M. | | 登録日 | 2015-03-12 | | 公開日 | 2015-05-27 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural mechanisms of DREAM complex assembly and regulation.
Genes Dev., 29, 2015
|
|
4YBU
 
 | |
4YCH
 
 | |
470D
 
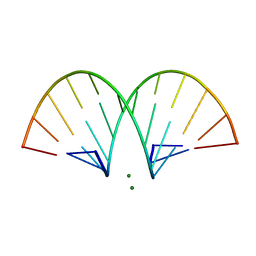 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | 分子名称: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | 著者 | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | 登録日 | 1999-04-29 | | 公開日 | 1999-05-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
5KM3
 
 | |
7QGS
 
 | | Crystal structure of p300 CH1 domain in complex with CITIF (a CITED2-HIF-1alpha hybrid) | | 分子名称: | Cbp/p300-interacting transactivator 2,Hypoxia-inducible factor 1-alpha, Histone acetyltransferase, ZINC ION | | 著者 | Hegedus, Z, Wilson, A.J, Edwards, T.A. | | 登録日 | 2021-12-10 | | 公開日 | 2022-05-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Understanding p300-transcription factor interactions using sequence variation and hybridization.
Rsc Chem Biol, 3, 2022
|
|
5KM1
 
 | |
5KMA
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside D-Trp phosphoramidate substrate complex | | 分子名称: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-ethyl-phosphonamidic acid | | 著者 | Maize, K.M, Finzel, B.C. | | 登録日 | 2016-06-26 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
3OGT
 
 | | Design, Chemical synthesis, Functional characterization and Crystal structure of the sidechain analogue of 1,25-dihydroxyvitamin D3. | | 分子名称: | (1S,3R,5Z,7E,14beta,17alpha,20S)-20-[5-(1-hydroxy-1-methylethyl)furan-2-yl]-9,10-secopregna-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | 著者 | Huet, T, Fraga, R, Mourino, A, Moras, D, Rochel, N. | | 登録日 | 2010-08-17 | | 公開日 | 2011-08-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Design, Chemical synthesis, Functional characterization and Crystal structure of the sidechain analogue of 1,25-dihydroxyvitamin D3.
To be Published
|
|
7QSM
 
 | | Bovine complex I in lipid nanodisc, Deactive-ligand (composite) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Chung, I, Bridges, H.R, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QSL
 
 | | Bovine complex I in lipid nanodisc, Active-apo | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Chung, I, Bridges, H.R, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
5KTX
 
 | | CREBBP bromodomain in complex with Cpd59 ((S)-1-(3-((2-fluoro-4-(1-methyl-1H-pyrazol-4-yl)phenyl)amino)-1-(tetrahydrofuran-3-yl)-6,7-dihydro-1H-pyrazolo[4,3-c]pyridin-5(4H)-yl)ethanone) | | 分子名称: | 1,2-ETHANEDIOL, 1-[3-[[2-fluoranyl-4-(1-methylpyrazol-4-yl)phenyl]amino]-1-[(3~{S})-oxolan-3-yl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-5-yl]ethanone, CREB-binding protein, ... | | 著者 | Murray, J.M, Noland, C. | | 登録日 | 2016-07-12 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
7QSN
 
 | | Bovine complex I in lipid nanodisc, Deactive-apo | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Chung, I, Bridges, H.R, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QSK
 
 | | Bovine complex I in lipid nanodisc, Active-Q10 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Chung, I, Bridges, H.R, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QSO
 
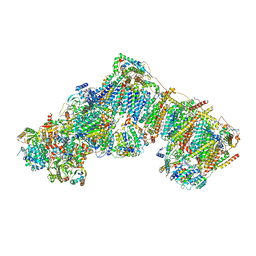 | | Bovine complex I in lipid nanodisc, State 3 (Slack) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Chung, I, Bridges, H.R, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
3FWB
 
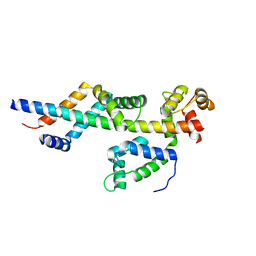 | | Sac3:Sus1:Cdc31 complex | | 分子名称: | Cell division control protein 31, Nuclear mRNA export protein SAC3, Protein SUS1 | | 著者 | Stewart, M, Jani, D. | | 登録日 | 2009-01-17 | | 公開日 | 2009-04-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export.
Mol.Cell, 33, 2009
|
|
4YXP
 
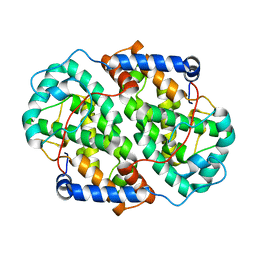 | | The structure of the folded domain of the signature multifunctional protein ICP27 from herpes simplex virus-1 reveals an intertwined dimer. | | 分子名称: | ZINC ION, mRNA export factor | | 著者 | Tunnicliffe, R.B, Schacht, M, Levy, C.W, Jowitt, T.A, Sandri-Goldin, R.M, Golovanov, A.P. | | 登録日 | 2015-03-23 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | The structure of the folded domain from the signature multifunctional protein ICP27 from herpes simplex virus-1 reveals an intertwined dimer.
Sci Rep, 5, 2015
|
|
8V84
 
 | | 60S ribosome biogenesis intermediate (Dbp10 catalytic structure - Overall map) | | 分子名称: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | 著者 | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | 登録日 | 2023-12-04 | | 公開日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8V83
 
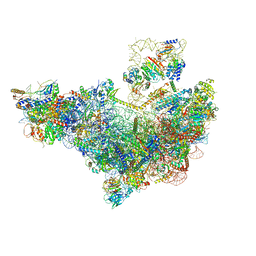 | | 60S ribosome biogenesis intermediate (Dbp10 pre-catalytic structure - Overall map) | | 分子名称: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | 著者 | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | 登録日 | 2023-12-04 | | 公開日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.53 Å) | | 主引用文献 | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
5IQL
 
 | |
4Z5R
 
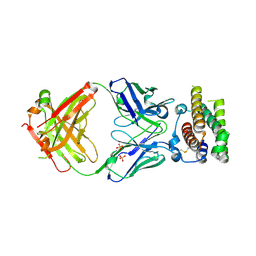 | | Rontalizumab Fab bound to Interferon-a2 | | 分子名称: | Interferon alpha-2, SULFATE ION, anti-IFN-a antibody rontalizumab heavy chain modules VH and CH1 (Fab), ... | | 著者 | Eigenbrot, C, Maurer, B, Bosanac, I. | | 登録日 | 2015-04-02 | | 公開日 | 2015-07-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis of the broadly neutralizing anti-interferon-alpha antibody rontalizumab.
Protein Sci., 24, 2015
|
|
6Q7U
 
 | |
5IOK
 
 | |
