5X7X
 
 | | The crystal structure of the nucleosome containing H3.3 at 2.18 angstrom resolution | | 分子名称: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | 著者 | Arimura, Y, Taguchi, H, Kurumizaka, H. | | 登録日 | 2017-02-27 | | 公開日 | 2017-04-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.184 Å) | | 主引用文献 | Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8
Biochemistry, 56, 2017
|
|
2I5J
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with DHBNH, an RNASE H inhibitor | | 分子名称: | (E)-3,4-DIHYDROXY-N'-[(2-METHOXYNAPHTHALEN-1-YL)METHYLENE]BENZOHYDRAZIDE, MAGNESIUM ION, Reverse transcriptase/ribonuclease H P51 subunit, ... | | 著者 | Himmel, D.M, Sarafianos, S.G, Knight, J.L, Levy, R.M, Arnold, E. | | 登録日 | 2006-08-24 | | 公開日 | 2006-12-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | HIV-1 reverse transcriptase structure with RNase H inhibitor dihydroxy benzoyl naphthyl hydrazone bound at a novel site.
Acs Chem.Biol., 1, 2006
|
|
5K14
 
 | | HIV-1 Reverse Transcriptase in complex with a 2,6-difluorophenyl DAPY analog | | 分子名称: | 4-{[4-(2,6-difluoro-4-methoxybenzene-1-carbonyl)pyrimidin-2-yl]amino}benzonitrile, HIV-1 reverse transcriptase (isolate LW123), HIV-1 reverse transcriptase(isolate HXB2) | | 著者 | Lansdon, E.B. | | 登録日 | 2016-05-17 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.402 Å) | | 主引用文献 | Novel (2,6-difluorophenyl)(2-(phenylamino)pyrimidin-4-yl)methanones with restricted conformation as potent non-nucleoside reverse transcriptase inhibitors against HIV-1.
Eur.J.Med.Chem., 122, 2016
|
|
3CTM
 
 | | Crystal Structure of a Carbonyl Reductase from Candida Parapsilosis with anti-Prelog Stereo-specificity | | 分子名称: | Carbonyl Reductase | | 著者 | Zhang, R, Zhu, G, Li, X, Xu, Y, Zhang, X.C, Rao, Z. | | 登録日 | 2008-04-14 | | 公開日 | 2008-05-27 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Crystal structure of a carbonyl reductase from Candida parapsilosis with anti-Prelog stereospecificity.
Protein Sci., 17, 2008
|
|
5XM1
 
 | | The mouse nucleosome structure containing H2A, H2B type3-A, H3mm7, and H4 | | 分子名称: | DNA (146-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | 著者 | Taguchi, H, Horikoshi, N, Kurumizaka, H. | | 登録日 | 2017-05-12 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration.
Nat Commun, 9, 2018
|
|
8SVF
 
 | | BAP1/ASXL1 bound to the H2AK119Ub Nucleosome | | 分子名称: | DNA/RNA (187-MER), DNA/RNA (327-MER), Histone H2A type 1, ... | | 著者 | Thomas, J.F, Valencia-Sanchez, M.I, Armache, K.-J. | | 登録日 | 2023-05-16 | | 公開日 | 2023-08-30 | | 最終更新日 | 2024-11-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of histone H2A lysine 119 deubiquitination by Polycomb repressive deubiquitinase BAP1/ASXL1.
Sci Adv, 9, 2023
|
|
6O19
 
 | | Crystal Structure of Pho7 complex with pho1 promoter site 2 | | 分子名称: | DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*AP*TP*TP*TP*CP*CP*GP*AP*AP*TP*AP*AP*T)-3'), DNA (5'-D(*TP*TP*AP*TP*TP*CP*GP*GP*AP*AP*AP*TP*TP*AP*AP*AP*AP*AP*CP*A)-3'), Transcription factor Pho7, ... | | 著者 | Garg, A, Goldgur, Y, Shuman, S. | | 登録日 | 2019-02-18 | | 公開日 | 2019-04-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.596 Å) | | 主引用文献 | Structure of Fission Yeast Transcription Factor Pho7 Bound topho1Promoter DNA and Effect of Pho7 Mutations on DNA Binding and Phosphate Homeostasis.
Mol.Cell.Biol., 39, 2019
|
|
1VHW
 
 | |
1VHJ
 
 | |
2WVJ
 
 | | Mutation of Thr163 to Ser in Human Thymidine Kinase Shifts the Specificity from Thymidine towards the Nucleoside Analogue Azidothymidine | | 分子名称: | MAGNESIUM ION, THYMIDINE KINASE, CYTOSOLIC, ... | | 著者 | Skovgaard, T, Uhlin, U, Munch-Petersen, B. | | 登録日 | 2009-10-17 | | 公開日 | 2009-10-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Comparative Active-Site Mutation Study of Human and Caenorhabditis Elegans Thymidine Kinase 1.
FEBS J., 279, 2012
|
|
3UAZ
 
 | | Crystal structure of Bacillus cereus adenosine phosphorylase D204N mutant complexed with inosine | | 分子名称: | GLYCEROL, INOSINE, Purine nucleoside phosphorylase deoD-type, ... | | 著者 | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | 登録日 | 2011-10-22 | | 公開日 | 2012-02-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3UAY
 
 | | Crystal structure of Bacillus cereus adenosine phosphorylase D204N mutant complexed with adenosine | | 分子名称: | ADENOSINE, GLYCEROL, Purine nucleoside phosphorylase deoD-type, ... | | 著者 | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | 登録日 | 2011-10-22 | | 公開日 | 2012-02-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3UAW
 
 | | Crystal structure of adenosine phosphorylase from Bacillus cereus complexed with adenosine | | 分子名称: | ADENOSINE, GLYCEROL, Purine nucleoside phosphorylase deoD-type, ... | | 著者 | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | 登録日 | 2011-10-22 | | 公開日 | 2012-02-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8HAL
 
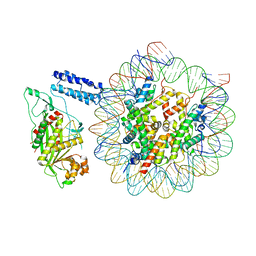 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 1 | | 分子名称: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAG
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 1 (3.2 angstrom resolution) | | 分子名称: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAM
 
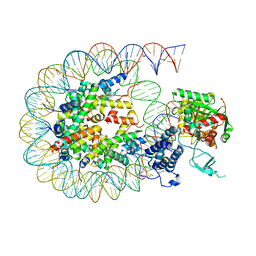 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 2 | | 分子名称: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAH
 
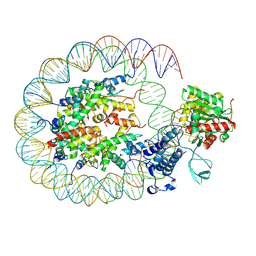 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 2 (3.9 angstrom resolution) | | 分子名称: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAI
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 1 (4.7 angstrom resolution) | | 分子名称: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAN
 
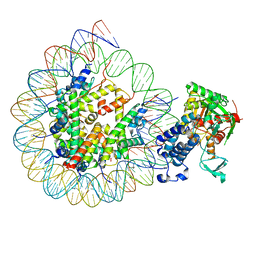 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 3 | | 分子名称: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAJ
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 2 (4.8 angstrom resolution) | | 分子名称: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAK
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 4 (4.5 angstrom resolution) | | 分子名称: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | 登録日 | 2022-10-26 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
3UAV
 
 | | Crystal structure of adenosine phosphorylase from Bacillus cereus | | 分子名称: | GLYCEROL, Purine nucleoside phosphorylase deoD-type, SULFATE ION | | 著者 | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | 登録日 | 2011-10-22 | | 公開日 | 2012-02-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3UAX
 
 | | Crystal structure of adenosine phosphorylase from Bacillus cereus complexed with inosine | | 分子名称: | GLYCEROL, INOSINE, Purine nucleoside phosphorylase deoD-type, ... | | 著者 | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | 登録日 | 2011-10-22 | | 公開日 | 2012-02-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1UIO
 
 | | ADENOSINE DEAMINASE (HIS 238 ALA MUTANT) | | 分子名称: | 6-HYDROXY-7,8-DIHYDRO PURINE NUCLEOSIDE, ADENOSINE DEAMINASE, ZINC ION | | 著者 | Wilson, D.K, Quiocho, F.A. | | 登録日 | 1996-08-30 | | 公開日 | 1997-06-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Site-directed mutagenesis of histidine 238 in mouse adenosine deaminase: substitution of histidine 238 does not impede hydroxylate formation.
Biochemistry, 35, 1996
|
|
1RT3
 
 | | AZT DRUG RESISTANT HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH 1051U91 | | 分子名称: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE | | 著者 | Ren, J, Stammers, D.K, Stuart, D.I. | | 登録日 | 1998-06-29 | | 公開日 | 1999-02-16 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | 3'-Azido-3'-deoxythymidine drug resistance mutations in HIV-1 reverse transcriptase can induce long range conformational changes.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
