3GY8
 
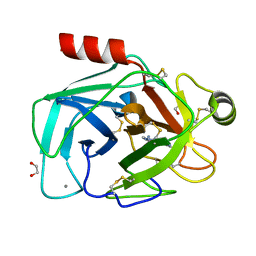 | | A comparative study on the inhibition of bovine beta-trypsin by bis-benzamidines diminazene and pentamidine by X-ray crystallography and ITC | | 分子名称: | 1,2-ETHANEDIOL, BERENIL, CALCIUM ION, ... | | 著者 | Perilo, C.S, Pereira, M.T, Santoro, M.M, Nagem, R.A.P. | | 登録日 | 2009-04-03 | | 公開日 | 2010-03-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural binding evidence of the trypanocidal drugs Berenil and Pentacarinate active principles to a serine protease model.
Int.J.Biol.Macromol., 46, 2010
|
|
7SKI
 
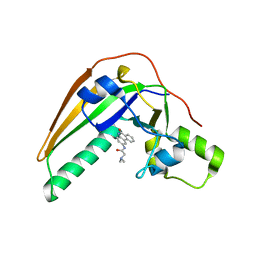 | | Pertussis toxin in complex with PJ34 | | 分子名称: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Pertussis toxin subunit 1 | | 著者 | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | 登録日 | 2021-10-20 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.09912443 Å) | | 主引用文献 | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
8OO9
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-DNA refinement state1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase INO80, DNA strand 1, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-04 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
2G19
 
 | |
5K4X
 
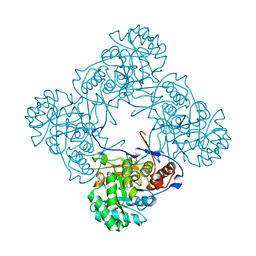 | | M. thermoresistible IMPDH in complex with IMP and Compound 1 | | 分子名称: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ~{N}-(2~{H}-indazol-6-yl)-3,5-dimethyl-1~{H}-pyrazole-4-sulfonamide | | 著者 | Pacitto, A, Ascher, D.B, Blundell, T.L. | | 登録日 | 2016-05-22 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis.
ACS Infect Dis, 3, 2017
|
|
2YS9
 
 | | structure of the third Homeodomain from the human homeobox and leucine zipper protein, Homez | | 分子名称: | Homeobox and leucine zipper protein Homez | | 著者 | Ohnishi, S, Tomizawa, T, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-03 | | 公開日 | 2007-10-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | structure of the third Homeodomain from the human homeobox and leucine zipper protein, Homez
To be Published
|
|
8OOF
 
 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state1 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOT
 
 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state2 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
2G1M
 
 | |
5K54
 
 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state | | 分子名称: | Fructose-1,6-bisphosphatase isozyme 2 | | 著者 | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | 登録日 | 2016-05-23 | | 公開日 | 2017-06-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.717 Å) | | 主引用文献 | Structural studies of human muscle FBPase
To Be Published
|
|
2LFH
 
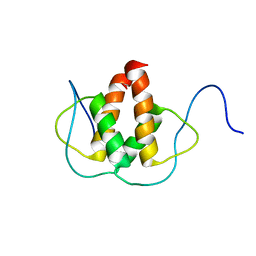 | | Solution NMR Structure of the Helix-loop-Helix Domain of Human ID3 Protein, Northeast Structural Genomics Consortium Target HR3111A | | 分子名称: | DNA-binding protein inhibitor ID-3 | | 著者 | Eletsky, A, Wang, D, Kohan, E, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-06-30 | | 公開日 | 2011-07-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of the Helix-loop-Helix Domain of Human ID3 Protein, Northeast Structural Genomics Consortium Target HR3111A
To be Published
|
|
7DCU
 
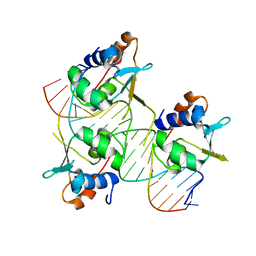 | |
3H7Z
 
 | |
3H0M
 
 | | Structure of trna-dependent amidotransferase gatcab from aquifex aeolicus | | 分子名称: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, GLUTAMINE, Glutamyl-tRNA(Gln) amidotransferase subunit A, ... | | 著者 | Wu, J, Bu, W, Sheppard, K, Kitabatake, M, Soll, D, Smith, J.L. | | 登録日 | 2009-04-09 | | 公開日 | 2009-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Insights into tRNA-Dependent Amidotransferase Evolution and Catalysis from the Structure of the Aquifex aeolicus Enzyme
J.Mol.Biol., 391, 2009
|
|
7RZW
 
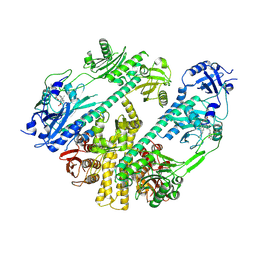 | | CryoEM structure of Arabidopsis thaliana phytochrome B | | 分子名称: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B | | 著者 | Li, H, Burgie, E.S, Vierstra, R.D, Li, H. | | 登録日 | 2021-08-27 | | 公開日 | 2022-04-13 | | 最終更新日 | 2022-04-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Plant phytochrome B is an asymmetric dimer with unique signalling potential.
Nature, 604, 2022
|
|
2YTN
 
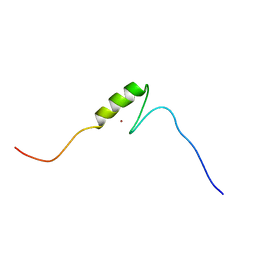 | | Solution structure of the C2H2 type zinc finger (region 732-764) of human Zinc finger protein 347 | | 分子名称: | ZINC ION, Zinc finger protein 347 | | 著者 | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-05 | | 公開日 | 2007-10-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the C2H2 type zinc finger (region 732-764) of human Zinc finger protein 347
To be Published
|
|
7DDE
 
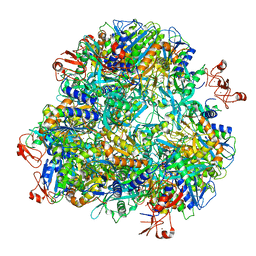 | | Cryo-EM structure of the Ape4 and Nbr1 complex | | 分子名称: | Aspartyl aminopeptidase 1,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | 著者 | Zhang, J, Ye, K. | | 登録日 | 2020-10-28 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.26 Å) | | 主引用文献 | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
2YU1
 
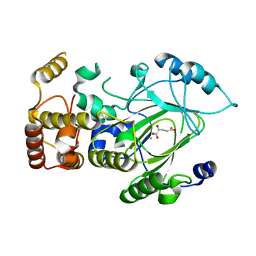 | |
7OLA
 
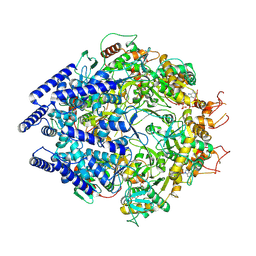 | | Structure of Primase-Helicase in SaPI5 | | 分子名称: | DNA primase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Qiao, C.C, Mir-Sanchis, I. | | 登録日 | 2021-05-19 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Staphylococcal self-loading helicases couple the staircase mechanism with inter domain high flexibility.
Nucleic Acids Res., 50, 2022
|
|
2LFB
 
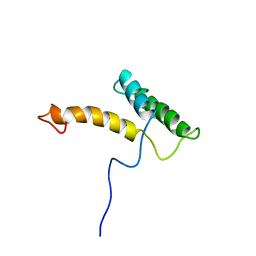 | | HOMEODOMAIN FROM RAT LIVER LFB1/HNF1 TRANSCRIPTION FACTOR, NMR, 20 STRUCTURES | | 分子名称: | LFB1/HNF1 TRANSCRIPTION FACTOR | | 著者 | Schott, O, Billeter, M, Leiting, B, Wider, G, Wuthrich, K. | | 登録日 | 1996-12-12 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The NMR solution structure of the non-classical homeodomain from the rat liver LFB1/HNF1 transcription factor.
J.Mol.Biol., 267, 1997
|
|
5JRJ
 
 | | Crystal Structure of Herbaspirillum seropedicae RecA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | 著者 | Leite, W.C, Galvao, C.W, Saab, S.C, Iulek, J, Etto, R.M, Steffens, M.B.R, Chitteni-Pattu, S, Stanage, T, Keck, J.L, Cox, M.M. | | 登録日 | 2016-05-06 | | 公開日 | 2016-08-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and Functional Studies of H. seropedicae RecA Protein - Insights into the Polymerization of RecA Protein as Nucleoprotein Filament.
PLoS ONE, 11, 2016
|
|
7PBQ
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0+A [t2 dataset] | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | 著者 | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | 登録日 | 2021-08-02 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
3H3W
 
 | | Fitting of the gp6 crystal structure into 3D cryo-EM reconstruction of bacteriophage T4 dome-shaped baseplate | | 分子名称: | Baseplate structural protein Gp6 | | 著者 | Aksyuk, A.A, Leiman, P.G, Shneider, M.M, Mesyanzhinov, V.V, Rossmann, M.G. | | 登録日 | 2009-04-17 | | 公開日 | 2009-05-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (12 Å) | | 主引用文献 | The structure of gene product 6 of bacteriophage T4, the hinge-pin of the baseplate.
Structure, 17, 2009
|
|
2YUW
 
 | |
7DD9
 
 | | Cryo-EM structure of the Ams1 and Nbr1 complex | | 分子名称: | Alpha-mannosidase,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | 著者 | Zhang, J, Ye, K. | | 登録日 | 2020-10-28 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
