6IK5
 
 | | Crystal structure of tomato beta-galactosidase (TBG) 4 in complex with galactose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | 著者 | Matsuyama, K, Nakae, S, Igarashi, K, Tada, T, Ishimaru, M. | | 登録日 | 2018-10-15 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Substrate-recognition mechanism of tomato beta-galactosidase 4 using X-ray crystallography and docking simulation.
Planta, 252, 2020
|
|
5XE1
 
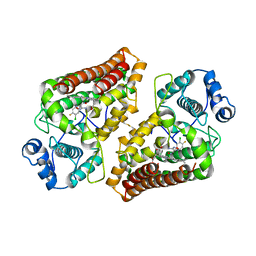 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with INCB14943 | | 分子名称: | 4-Amino-N-(3-chloro-4-fluorophenyl)-N'-hydroxy-1,2,5-oxadiazole-3-carboxamidine, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Xu, J, Wu, U, Liu, J. | | 登録日 | 2017-03-31 | | 公開日 | 2017-05-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural insights into the binding mechanism of IDO1 with hydroxylamidine based inhibitor INCB14943
Biochem. Biophys. Res. Commun., 487, 2017
|
|
3DEC
 
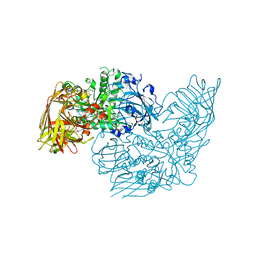 | | Crystal structure of a glycosyl hydrolases family 2 protein from Bacteroides thetaiotaomicron | | 分子名称: | Beta-galactosidase, POTASSIUM ION | | 著者 | Kumaran, D, Bonanno, J, Romero, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-06-09 | | 公開日 | 2008-06-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of a Glycosyl Hydrolases Family 2 protein from Bacteroides thetaiotaomicron.
To be Published
|
|
6ILI
 
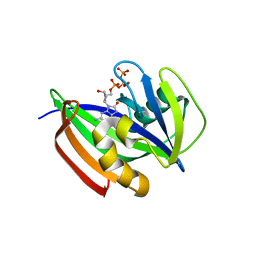 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 8-oxo-dGTP at pH 6.5 | | 分子名称: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE | | 著者 | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | 登録日 | 2018-10-18 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
4OGF
 
 | |
4K8D
 
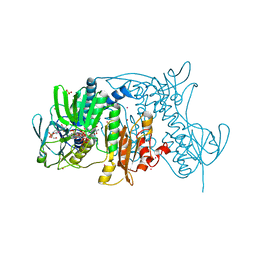 | | Crystal structure of the C558(464)A/C559(465)A double mutant of Tn501 MerA in complex with NADPH and Hg2+ | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Mercuric reductase, ... | | 著者 | Dong, A, Falkowski, M, Malone, M, Miller, S.M, Pai, E.F. | | 登録日 | 2013-04-18 | | 公開日 | 2013-05-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Crystal structure of the C136(42)A/C141(47)A double mutant of Tn501 MerA in complex with NADPH and
Hg2+
to be published
|
|
6DPQ
 
 | | Mapping the binding trajectory of a suicide inhibitor in human indoleamine 2,3-dioxygenase 1 | | 分子名称: | (2R)-N-(4-chlorophenyl)-2-[cis-4-(6-fluoroquinolin-4-yl)cyclohexyl]propanamide, GLYCEROL, Indoleamine 2,3-dioxygenase 1, ... | | 著者 | Pham, K.N, Yeh, S.R. | | 登録日 | 2018-06-09 | | 公開日 | 2018-11-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Mapping the Binding Trajectory of a Suicide Inhibitor in Human Indoleamine 2,3-Dioxygenase 1.
J. Am. Chem. Soc., 140, 2018
|
|
3DJY
 
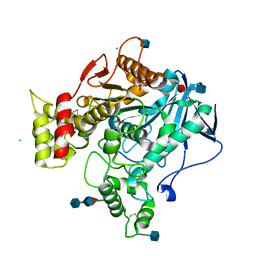 | | Nonaged Form of Human Butyrylcholinesterase Inhibited by Tabun | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Carletti, E, Nachon, F. | | 登録日 | 2008-06-24 | | 公開日 | 2008-12-02 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Aging of Cholinesterases Phosphylated by Tabun Proceeds through O-Dealkylation.
J.Am.Chem.Soc., 130, 2008
|
|
7PJV
 
 | | Structure of the 70S-EF-G-GDP-Pi ribosome complex with tRNAs in hybrid state 1 (H1-EF-G-GDP-Pi) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | 登録日 | 2021-08-24 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
4OGZ
 
 | |
4KDF
 
 | |
3GMZ
 
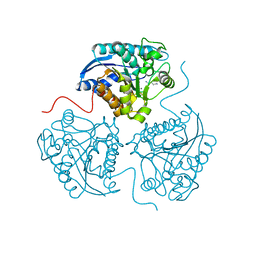 | |
7PJT
 
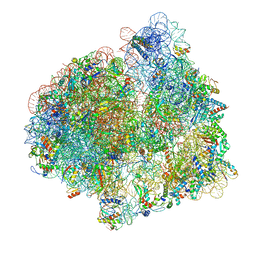 | | Structure of the 70S ribosome with tRNAs in hybrid state 1 (H1) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | 登録日 | 2021-08-24 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
6DQ3
 
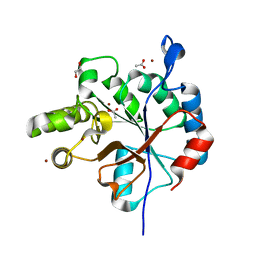 | | Streptococcus pyogenes deacetylase PplD in complex with acetate | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, IMIDAZOLE, ... | | 著者 | Li, J, Korotkova, N, Korotkov, K.V. | | 登録日 | 2018-06-10 | | 公開日 | 2019-05-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | PplD is a de-N-acetylase of the cell wall linkage unit of streptococcal rhamnopolysaccharides
Nat Commun, 13, 2022
|
|
3GO8
 
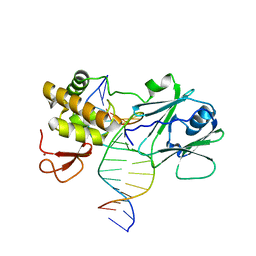 | | MutM encountering an intrahelical 8-oxoguanine (oxoG) lesion in EC3-loop deletion complex | | 分子名称: | 5'-D(*GP*CP*GP*TP*CP*CP*(8OG)P*GP*AP*TP*CP*TP*AP*C)-3', 5'-D(P*GP*GP*TP*AP*GP*AP*TP*CP*CP*GP*GP*AP*CP*G)-3', Formamidopyrimidine-DNA glycosylase, ... | | 著者 | Spong, M.C, Qi, Y, Verdine, G.L. | | 登録日 | 2009-03-18 | | 公開日 | 2009-12-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme
Nature, 462, 2009
|
|
3D9E
 
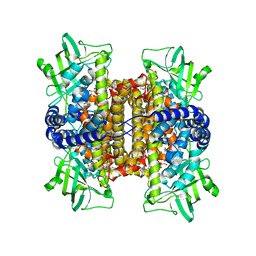 | | Nitroalkane oxidase: active site mutant D402N crystallized with 1-nitrooctane | | 分子名称: | 1-nitrooctane, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Heroux, A, Bozinovski, D.M, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | 登録日 | 2008-05-27 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of intermediates in the nitroalkane oxidase reaction.
Biochemistry, 48, 2009
|
|
7PJU
 
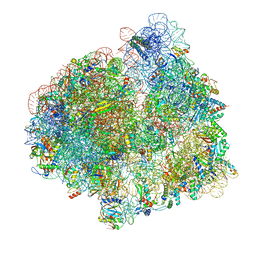 | | Structure of the 70S ribosome with tRNAs in hybrid state 2 (H2) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | 登録日 | 2021-08-24 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
6IQV
 
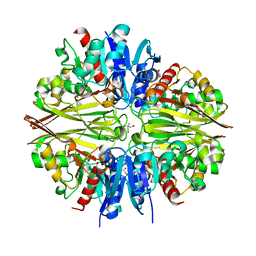 | | Crystal Structure of Cell Surface Glyceraldehyde-3-Phosphate Dehydrogenase Complexed with Hg2+ from Lactobacillus plantarum | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | 著者 | Yoneda, K, Kinoshita, H. | | 登録日 | 2018-11-09 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Crystal Structure of Cell Surface Glyceraldehyde-3-Phosphate Dehydrogenase from Lactobacillus plantarum: Insight into the Mercury Binding Mechanism
Milk Sci, 68, 2019
|
|
5ZZT
 
 | |
6IBM
 
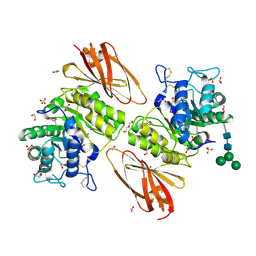 | | Crystal structure of human alpha-galactosidase A in complex with alpha-galactose configured cyclosulfate ME776 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | 著者 | Rowland, R.J, Wu, L, Davies, G.J. | | 登録日 | 2018-11-30 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Alpha-d-Gal-cyclophellitol cyclosulfamidate is a Michaelis complex analog that stabilizes therapeutic lysosomal alpha-galactosidase A in Fabry disease
Chem Sci, 2019
|
|
5ZZZ
 
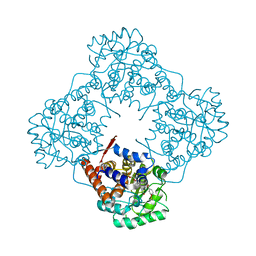 | |
4OMF
 
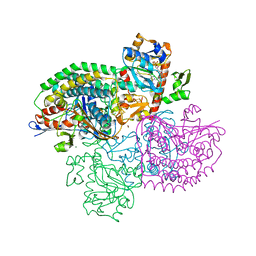 | | The F420-reducing [NiFe]-hydrogenase complex from Methanothermobacter marburgensis, the first X-ray structure of a group 3 family member | | 分子名称: | CHLORIDE ION, F420-reducing hydrogenase, subunit alpha, ... | | 著者 | Vitt, S, Ma, K, Warkentin, E, Moll, J, Pierik, A, Shima, S, Ermler, U. | | 登録日 | 2014-01-27 | | 公開日 | 2014-06-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | The F420-Reducing [NiFe]-Hydrogenase Complex from Methanothermobacter marburgensis, the First X-ray Structure of a Group 3 Family Member.
J.Mol.Biol., 426, 2014
|
|
4KF2
 
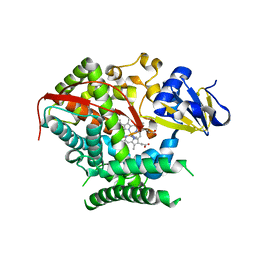 | | Structure of the P4509 BM3 A82F F87V heme domain | | 分子名称: | Bifunctional P-450/NADPH-P450 reductase, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Leys, D. | | 登録日 | 2013-04-26 | | 公開日 | 2013-07-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Key Mutations Alter the Cytochrome P450 BM3 Conformational Landscape and Remove Inherent Substrate Bias.
J.Biol.Chem., 288, 2013
|
|
6DUM
 
 | | ALDH1A1 N121S in complex with 6-{[(3-fluorophenyl)methyl]sulfanyl}-2-(oxetan-3-yl)-5-phenyl-2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one (compound 13g) | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 6-{[(3-fluorophenyl)methyl]sulfanyl}-2-(oxetan-3-yl)-5-phenyl-2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, ... | | 著者 | Buchman, C.D, Hurley, T.D. | | 登録日 | 2018-06-21 | | 公開日 | 2019-05-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Based Optimization of a Novel Class of Aldehyde Dehydrogenase 1A (ALDH1A) Subfamily-Selective Inhibitors as Potential Adjuncts to Ovarian Cancer Chemotherapy.
J.Med.Chem., 61, 2018
|
|
3GUK
 
 | |
