3CRI
 
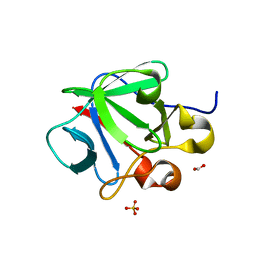 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ser, Glu82Asn and Lys101Ala | | 分子名称: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | 著者 | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | 登録日 | 2008-04-07 | | 公開日 | 2009-02-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3CRH
 
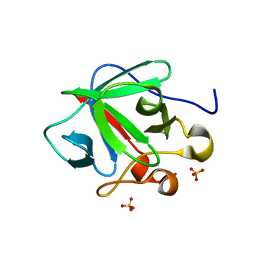 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ser and Lys101Ala | | 分子名称: | Heparin-binding growth factor 1, SULFATE ION | | 著者 | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | 登録日 | 2008-04-07 | | 公開日 | 2009-02-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3KK6
 
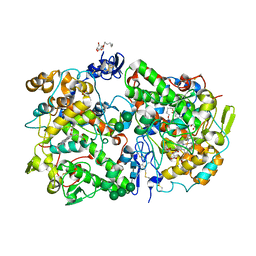 | | Crystal Structure of Cyclooxygenase-1 in complex with celecoxib | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[5-(4-METHYLPHENYL)-3-(TRIFLUOROMETHYL)-1H-PYRAZOL-1-YL]BENZENESULFONAMIDE, CITRATE ANION, ... | | 著者 | Sidhu, R.S. | | 登録日 | 2009-11-04 | | 公開日 | 2009-12-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Coxibs interfere with the action of aspirin by binding tightly to one monomer of cyclooxygenase-1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2KNX
 
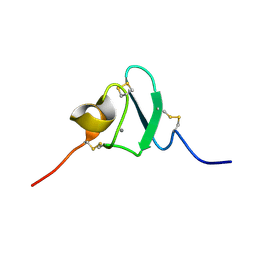 | |
1YQV
 
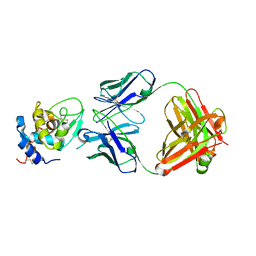 | | The crystal structure of the antibody Fab HyHEL5 complex with lysozyme at 1.7A resolution | | 分子名称: | Hen Egg White Lysozyme, HyHEL-5 Antibody Heavy Chain, HyHEL-5 Antibody Light Chain | | 著者 | Cohen, G.H, Silverton, E.W, Padlan, E.A, Dyda, F, Wibbenmeyer, J.A, Wilson, R.C, Davies, D.R. | | 登録日 | 2005-02-02 | | 公開日 | 2005-04-26 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Water molecules in the antibody-antigen interface of the structure of the Fab HyHEL-5-lysozyme complex at 1.7 A resolution: comparison with results from isothermal titration calorimetry.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4H9K
 
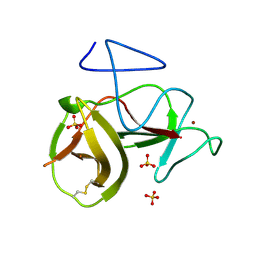 | | Crystal structure of cleavage site mutant of Npro of classical swine fever virus. | | 分子名称: | Hog cholera virus, SULFATE ION, ZINC ION | | 著者 | Gottipati, K, Ruggli, N, Gerber, M, Tratschin, J.-D, Benning, M, Bellamy, H, Choi, K.H. | | 登録日 | 2012-09-24 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | The Structure of Classical Swine Fever Virus N(pro): A Novel Cysteine Autoprotease and Zinc-Binding Protein Involved in Subversion of Type I Interferon Induction.
Plos Pathog., 9, 2013
|
|
3F9E
 
 | |
2KJD
 
 | |
5AOO
 
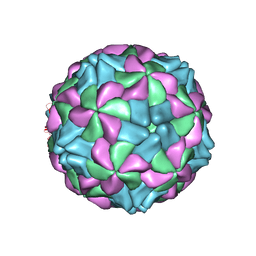 | | X-ray structure of a human Kobuvirus: Aichi virus A (AiV) | | 分子名称: | VP0, VP1, VP3 | | 著者 | Sabin, C, Palkova, L, Plevka, P. | | 登録日 | 2015-09-11 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Use of Noncrystallographic Symmetry Averaging to Solve Structures from Data Affected by Perfect Hemihedral Twinning
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5B2G
 
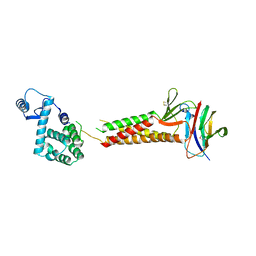 | | Crystal structure of human claudin-4 in complex with C-terminal fragment of Clostridium perfringens enterotoxin | | 分子名称: | Endolysin,Claudin-4, Heat-labile enterotoxin B chain | | 著者 | Shinoda, T, Kimura-Someya, T, Shirouzu, M, Yokoyama, S. | | 登録日 | 2016-01-15 | | 公開日 | 2016-10-05 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural basis for disruption of claudin assembly in tight junctions by an enterotoxin
Sci Rep, 6, 2016
|
|
4HXQ
 
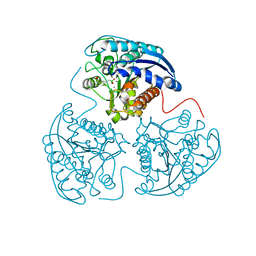 | | Crystal structure of human Arginase-1 complexed with inhibitor 14 | | 分子名称: | Arginase-1, MANGANESE (II) ION, [(5R)-5-carboxy-5-(methylamino)-7-(piperidin-1-yl)heptyl](trihydroxy)borate(1-) | | 著者 | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D.L, Golebiowski, A, Ji, M, Zhang, M, Beckett, P, Sheeler, R, Andreoli, M, Conway, B, Mahboubi, K, Schroeter, H, Van Zandt, M.C, Podjarny, A. | | 登録日 | 2012-11-12 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Discovery of (R)-2-Amino-6-borono-2-(2-(piperidin-1-yl)ethyl)hexanoic Acid and Congeners As Highly Potent Inhibitors of Human Arginases I and II for Treatment of Myocardial Reperfusion Injury.
J.Med.Chem., 56, 2013
|
|
4K6M
 
 | |
4LP8
 
 | | A Novel Open-State Crystal Structure of the Prokaryotic Inward Rectifier KirBac3.1 | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Inward rectifier potassium channel Kirbac3.1, ... | | 著者 | Zubcevic, L, Bavro, V.N, Muniz, J.R.C, Schmidt, M.R, Wang, S, De Zorzi, R, Venien-Bryan, C, Sansom, M.S.P, Nichols, C.G, Tucker, S.J. | | 登録日 | 2013-07-15 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Control of KirBac3.1 Potassium Channel Gating at the Interface between Cytoplasmic Domains.
J.Biol.Chem., 289, 2014
|
|
2GBV
 
 | | C6A/C111A/C57A/C146A holo CuZn Superoxide dismutase | | 分子名称: | COPPER (I) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | 著者 | Hornberg, A, Logan, D.T, Marklund, S.L, Oliveberg, M. | | 登録日 | 2006-03-11 | | 公開日 | 2007-01-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Coupling between Disulphide Status, Metallation and Dimer Interface Strength in Cu/Zn Superoxide Dismutase
J.Mol.Biol., 365, 2007
|
|
3CQA
 
 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ala and Lys101Ala | | 分子名称: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | 著者 | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | 登録日 | 2008-04-02 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1P63
 
 | | Human Acidic Fibroblast Growth Factor. 140 Amino Acid Form with Amino Terminal His Tag and Leu111 Replaced with Ile (L111I) | | 分子名称: | ACIDIC FIBROBLAST GROWTH FACTOR, FORMIC ACID, SULFATE ION | | 著者 | Brych, S.R, Kim, J, Logan, T.M, Blaber, M. | | 登録日 | 2003-04-28 | | 公開日 | 2004-05-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Accommodation of a highly symmetric core within a symmetric protein
superfold
Protein Sci., 12, 2003
|
|
4H9J
 
 | | Crystal structure of N-terminal protease (Npro) of classical swine fever virus. | | 分子名称: | Hog cholera virus | | 著者 | Gottipati, K, Ruggli, N, Gerber, M, Tratschin, J.-D, Benning, M, Bellamy, H, Choi, K.H. | | 登録日 | 2012-09-24 | | 公開日 | 2013-10-30 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The Structure of Classical Swine Fever Virus N(pro): A Novel Cysteine Autoprotease and Zinc-Binding Protein Involved in Subversion of Type I Interferon Induction.
Plos Pathog., 9, 2013
|
|
2KNY
 
 | |
3F9G
 
 | |
4NUG
 
 | |
4O1O
 
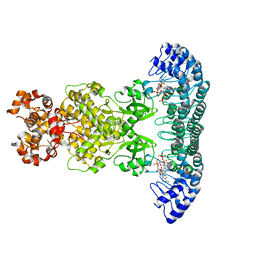 | | Crystal Structure of RNase L in complex with 2-5A | | 分子名称: | Ribonuclease L, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl] phosphono hydrogen phosphate | | 著者 | Huang, H, Zeqiraj, E, Ceccarelli, D.F, Sicheri, F. | | 登録日 | 2013-12-16 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.27 Å) | | 主引用文献 | Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity.
Mol.Cell, 53, 2014
|
|
4NUJ
 
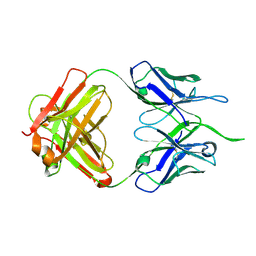 | |
4O1P
 
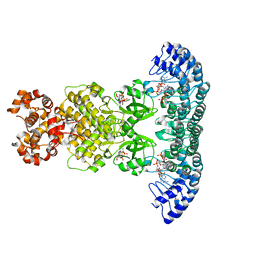 | | Crystal Structure of RNase L in complex with 2-5A and AMP-PNP | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribonuclease L, ... | | 著者 | Huang, H, Zeqiraj, E, Ceccarelli, D.F, Sicheri, F. | | 登録日 | 2013-12-16 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity.
Mol.Cell, 53, 2014
|
|
2R3E
 
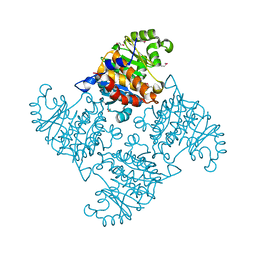 | |
2LUV
 
 | |
