6DXO
 
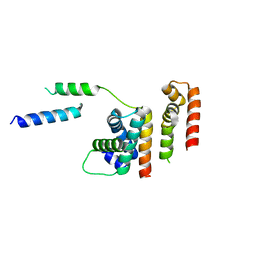 | | 1.8 A structure of RsbN-BldN complex. | | 分子名称: | BldN, RNA polymerase ECF-subfamily sigma factor | | 著者 | Schumacher, M.A. | | 登録日 | 2018-06-29 | | 公開日 | 2018-07-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structure of the RsbN-sigma BldN complex from Streptomyces venezuelae defines a new structural class of anti-sigma factor.
Nucleic Acids Res., 46, 2018
|
|
7M9J
 
 | | HIV-1 Protease WT (NL4-3) in Complex with LR3-68 | | 分子名称: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-4-[{4-[(1S)-1-hydroxyethyl]benzene-1-sulfonyl}(2-methylpropyl)amino]butyl}phenoxy)methyl]phosphonate | | 著者 | Lockbaum, G.J, Schiffer, C.A. | | 登録日 | 2021-03-31 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.859 Å) | | 主引用文献 | HIV-1 Protease in Complex with ligands
To Be Published
|
|
6DD7
 
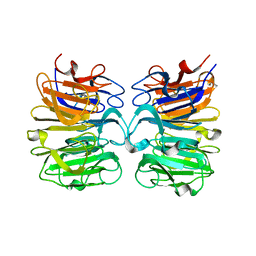 | |
8ECZ
 
 | | Bovine Fab 4C1 | | 分子名称: | 4C1 Fab heavy chain, 4C1 Fab light chain, PHOSPHATE ION | | 著者 | Stanfield, R.L, Wilson, I.A. | | 登録日 | 2022-09-02 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EDF
 
 | |
8ECQ
 
 | | Bovine Fab 2G3 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2G3 Fab Heavy chain, 2G3 Fab Light chain, ... | | 著者 | Stanfield, R.L, Wilson, I.A. | | 登録日 | 2022-09-02 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ECV
 
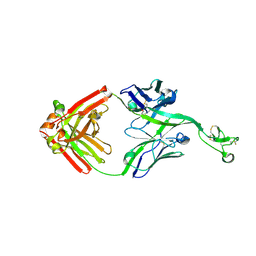 | | Bovine Fab 2F12 | | 分子名称: | 2F12 Fab Heavy chain, 2F12 Fab Light chain | | 著者 | Stanfield, R.L, Wilson, I.A. | | 登録日 | 2022-09-02 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ED1
 
 | | Bovine Fab 5C1 | | 分子名称: | 5C1 Fab heavy chain, 5C1 Fab light chain, GLYCEROL, ... | | 著者 | Stanfield, R.L, Wilson, I.A. | | 登録日 | 2022-09-02 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EW4
 
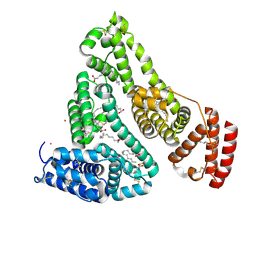 | | Human Serum Albumin with Cobalt (II) and Myristic Acid - crystal 1 | | 分子名称: | COBALT (II) ION, MYRISTIC ACID, Serum albumin | | 著者 | Gucwa, M, Cooper, D.R, Unciano, J, Lea, K, Kim, L, Lenkiewicz, J, Starban, I, Stewart, A.J, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-10-21 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and biochemical characterisation of Co2+-binding sites on serum albumins and their interplay with fatty acids
Chem Sci, 14, 2023
|
|
8EY5
 
 | | Human Serum Albumin with Cobalt (II) and Myristic Acid - crystal 3 | | 分子名称: | COBALT (II) ION, MYRISTIC ACID, Serum albumin | | 著者 | Gucwa, M, Cooper, D.R, Stewart, A.J, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-10-26 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural and biochemical characterisation of Co2+-binding sites on serum albumins and their interplay with fatty acids
Chem Sci, 14, 2023
|
|
8EW7
 
 | | Human Serum Albumin with Cobalt (II) and Myristic Acid - crystal 2 | | 分子名称: | COBALT (II) ION, MYRISTIC ACID, Serum albumin | | 著者 | Gucwa, M, Cooper, D.R, Stewart, A.J, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-10-21 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural and biochemical characterisation of Co2+-binding sites on serum albumins and their interplay with fatty acids
Chem Sci, 14, 2023
|
|
8EQ9
 
 | | Co-crystal structure of PERK with compound 11 | | 分子名称: | (2R)-N-[(4M)-4-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-3-methylphenyl]-2-(3-fluorophenyl)-2-hydroxyacetamide, Eukaryotic translation initiation factor 2-alpha kinase 3 | | 著者 | Zhu, G, Surman, M.D, Mulvihill, M.J. | | 登録日 | 2022-10-07 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Optimization of a Novel Mandelamide-Derived Pyrrolopyrimidine Series of PERK Inhibitors.
Pharmaceutics, 14, 2022
|
|
8EQD
 
 | | Co-crystal structure of PERK with compound 24 | | 分子名称: | (2R)-N-[(4M)-4-(4-amino-2,7-dimethyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-3-methylphenyl]-2-(3-fluorophenyl)-2-hydroxyacetamide, Eukaryotic translation initiation factor 2-alpha kinase 3 | | 著者 | Zhu, G, Surman, M.D, Mulvihill, M.J. | | 登録日 | 2022-10-07 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | Optimization of a Novel Mandelamide-Derived Pyrrolopyrimidine Series of PERK Inhibitors.
Pharmaceutics, 14, 2022
|
|
8EQE
 
 | | Co-crystal structure of PERK with compound 26 | | 分子名称: | (2R)-N-[(4M)-4-(4-amino-2,7-dimethyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-3-methylphenyl]-2-hydroxy-2-[3-(trifluoromethyl)phenyl]acetamide, Eukaryotic translation initiation factor 2-alpha kinase 3 | | 著者 | Zhu, G, Surman, M.D, Mulvihill, M.J. | | 登録日 | 2022-10-07 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.559 Å) | | 主引用文献 | Optimization of a Novel Mandelamide-Derived Pyrrolopyrimidine Series of PERK Inhibitors.
Pharmaceutics, 14, 2022
|
|
7NDV
 
 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001888. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[4-(trifluoromethyl)phenoxy]piperidine, Acetylcholine-binding protein, ... | | 著者 | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | 登録日 | 2021-02-02 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Discovery of fragments inducing conformational effects in dynamic proteins using a second-harmonic generation biosensor
RSC Advances, 11, 2021
|
|
7NDP
 
 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001856. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-bromanylspiro[3~{H}-chromene-2,4'-piperidine]-4-one, ... | | 著者 | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | 登録日 | 2021-02-02 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of fragments inducing conformational effects in dynamic proteins using a second-harmonic generation biosensor.
Rsc Adv, 11, 2021
|
|
7NN7
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with dimethyl 5-hydroxyisophthalate. | | 分子名称: | 1,2-ETHANEDIOL, Acetylglutamate kinase, SULFATE ION, ... | | 著者 | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | 登録日 | 2021-02-24 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.172 Å) | | 主引用文献 | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7N6V
 
 | |
7N6X
 
 | |
3ACL
 
 | | Crystal Structure of Human Pirin in complex with Triphenyl Compound | | 分子名称: | FE (II) ION, N-{[4-(benzyloxy)phenyl](methyl)-lambda~4~-sulfanylidene}-4-methylbenzenesulfonamide, Pirin | | 著者 | Okumura, H, Miyazaki, I, Simizu, S, Osada, H. | | 登録日 | 2010-01-05 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A small-molecule inhibitor shows that pirin regulates migration of melanoma cells
Nat.Chem.Biol., 6, 2010
|
|
7NG1
 
 | | Crystal structure of alpha Carbonic anhydrase from Schistoso ma mansoni bound to 1-(4-iodophenyl)-3-[2-(4-sulfamoylphenyl)ethyl]selenourea | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carbonic anhydrase, GLYCEROL, ... | | 著者 | Ferraroni, M, Angeli, A, Supuran, C.T. | | 登録日 | 2021-02-08 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Structural Insights into Schistosoma mansoni Carbonic Anhydrase (SmCA) Inhibition by Selenoureido-Substituted Benzenesulfonamides.
J.Med.Chem., 64, 2021
|
|
7NIY
 
 | | E. coli NfsA with FMN | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Day, M.D, Jarrom, D, Hyde, E.I, White, S.A. | | 登録日 | 2021-02-14 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
7NNX
 
 | | E. coli NfsA with 1,4-benzoquinone | | 分子名称: | 1,2-ETHANEDIOL, 1,4-benzoquinone, CHLORIDE ION, ... | | 著者 | Day, M.D, Jarrom, D, Hyde, E.I, White, S.A. | | 登録日 | 2021-02-25 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
7NMP
 
 | | E. coli NfsA with hydroquinone | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Day, M.D, Jarrom, D, Parr, R.J, Hyde, E.I, White, S.A. | | 登録日 | 2021-02-23 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
1Z1H
 
 | | HIV-1 protease complexed with macrocyclic peptidomimetic inhibitor 3 | | 分子名称: | N-{(2R)-2-HYDROXY-2-[(8S,11S)-8-ISOPROPYL-6,9-DIOXO-2-OXA-7,10-DIAZABICYCLO[11.2.2]HEPTADECA-1(15),13,16-TRIEN-11-YL]ETHYL}-N-ISOPENTYLBENZENESULFONAMIDE, Pol polyprotein, SULFATE ION | | 著者 | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | 登録日 | 2005-03-04 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease
Biochemistry, 38, 1999
|
|
