4YXF
 
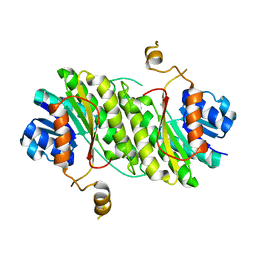 | |
4ZEL
 
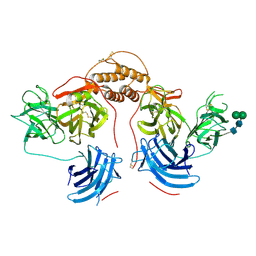 | | Human dopamine beta-hydroxylase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | 著者 | Vendelboe, T.V, Harris, P, Christensen, H.E.M, Harlos, K, Walter, T, Zhao, Y, Omari, K. | | 登録日 | 2015-04-20 | | 公開日 | 2016-04-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The crystal structure of human dopamine beta-hydroxylase at 2.9 angstrom resolution.
Sci Adv, 2, 2016
|
|
6ZO2
 
 | | 1.65 A resolution 4,5-dimethylcatechol (4,5-dimethylbenzene-1,2-diol) inhibited Sporosarcina pasteurii urease | | 分子名称: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | 著者 | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | 登録日 | 2020-07-07 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Inhibition of Urease, a Ni-Enzyme: The Reactivity of a Key Thiol With Mono- and Di-Substituted Catechols Elucidated by Kinetic, Structural, and Theoretical Studies.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
2OK6
 
 | |
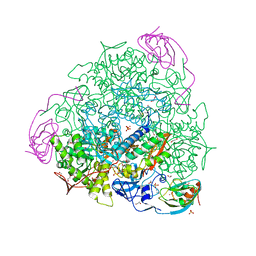
6ZR2
 
 | | Cryo-EM structure of respiratory complex I in the active state from Mus musculus at 3.1 A | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Bridges, H.R, Blaza, J.N, Agip, A.N.A, Hirst, J. | | 登録日 | 2020-07-10 | | 公開日 | 2020-10-21 | | 最終更新日 | 2020-10-28 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure of inhibitor-bound mammalian complex I.
Nat Commun, 11, 2020
|
|
4ZFM
 
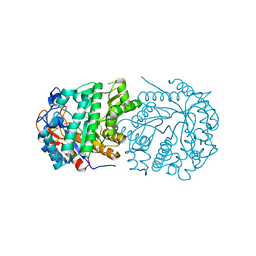 | | Structure of Gan1D-E170Q in complex with cellobiose-6-phosphate | | 分子名称: | 1,5-anhydro-6-O-phosphono-D-glucitol, 1,5-anhydro-6-O-phosphono-D-glucitol-(1-4)-beta-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, ... | | 著者 | Lansky, S, Zehavi, A, Dvir, H, Shoham, Y, Shoham, G. | | 登録日 | 2015-04-21 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure of Gan1D-E170Q in complex with cellobiose-6-phosphate
To Be Published
|
|
2ODN
 
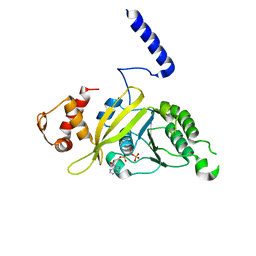 | | MSRECA-dATP complex | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Protein recA | | 著者 | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | 登録日 | 2006-12-24 | | 公開日 | 2007-06-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
2JE9
 
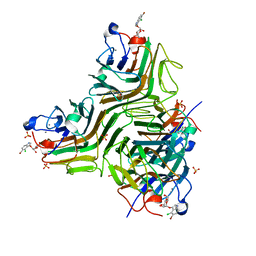 | | CRYSTAL STRUCTURE OF RECOMBINANT DIOCLEA GRANDIFLORA LECTIN COMPLEXED WITH 5-BROMO-4-CHLORO-3-INDOLYL-A-D-MANNOSE | | 分子名称: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, LECTIN ALPHA CHAIN, ... | | 著者 | Nagano, C.S, Sanz, L, Cavada, B.S, Calvete, J.J. | | 登録日 | 2007-01-16 | | 公開日 | 2007-10-30 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Insights Into the Structural Basis of the Ph- Dependent Dimer-Tetramer Equilibrium Through Crystallographic Analysis of Recombinant Diocleinae Lectins.
Biochem.J., 409, 2008
|
|
2OFI
 
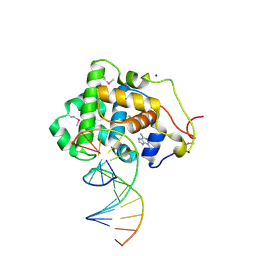 | | Crystal Structure of 3-methyladenine DNA Glycosylase I (TAG) bound to DNA/3mA | | 分子名称: | 3-METHYL-3H-PURIN-6-YLAMINE, 3-methyladenine DNA glycosylase I, constitutive, ... | | 著者 | Metz, A.H, Hollis, T, Eichman, B.F. | | 登録日 | 2007-01-03 | | 公開日 | 2007-05-15 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | DNA damage recognition and repair by 3-methyladenine DNA glycosylase I (TAG).
Embo J., 26, 2007
|
|
6TNZ
 
 | | Human polymerase delta-FEN1-PCNA toolbelt | | 分子名称: | DNA polymerase delta catalytic subunit, DNA polymerase delta subunit 2, DNA polymerase delta subunit 3, ... | | 著者 | Lancey, C, Hamdan, S.M, De Biasio, A. | | 登録日 | 2019-12-10 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Structure of the processive human Pol delta holoenzyme.
Nat Commun, 11, 2020
|
|
6ZS8
 
 | |
6TOZ
 
 | | Crystal structure of Bacillus paralicheniformis alpha-amylase in complex with acarbose | | 分子名称: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ACETIC ACID, Amylase, ... | | 著者 | Rozeboom, H.J, Janssen, D.B. | | 登録日 | 2019-12-12 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Characterization of the starch surface binding site on Bacillus paralicheniformis alpha-amylase.
Int.J.Biol.Macromol., 165, 2020
|
|
4Z3M
 
 | | X-ray structure of the adduct formed in the reaction between lysozyme and a platinum(II) Complex with S,O Bidentate Ligands (9b) | | 分子名称: | 1,2-ETHANEDIOL, 3-[2-chloranyl-2-[dimethyl(oxidanyl)-{4}-sulfanyl]-4-ethylsulfanyl-1-oxa-3{3}-thia-2{4}-platinacyclohexa-3,5-dien-6-yl]phenol, DIMETHYL SULFOXIDE, ... | | 著者 | Merlino, A. | | 登録日 | 2015-03-31 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Platinum(II) Complexes with O,S Bidentate Ligands: Biophysical Characterization, Antiproliferative Activity, and Crystallographic Evidence of Protein Binding.
Inorg.Chem., 54, 2015
|
|
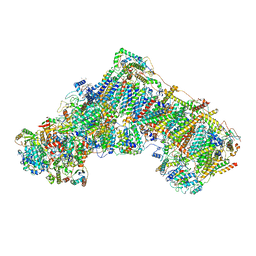
7AR7
 
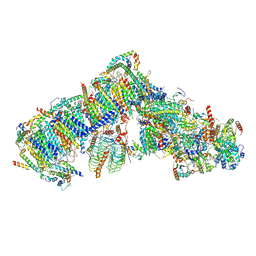 | | Cryo-EM structure of Arabidopsis thaliana complex-I (open conformation) | | 分子名称: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, ... | | 著者 | Klusch, N, Kuelbrandt, W. | | 登録日 | 2020-10-23 | | 公開日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.72 Å) | | 主引用文献 | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
4Z40
 
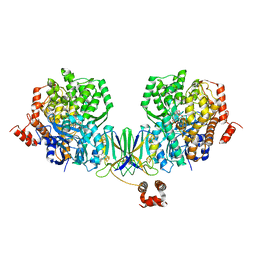 | | Active site complex BamBC of Benzoyl Coenzyme A reductase as isolated | | 分子名称: | Benzoyl-CoA reductase, putative, IRON/SULFUR CLUSTER, ... | | 著者 | Weinert, T, Kung, J.W, Weidenweber, S, Huwiler, S.G, Boll, M, Ermler, U. | | 登録日 | 2015-04-01 | | 公開日 | 2015-06-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural basis of enzymatic benzene ring reduction.
Nat.Chem.Biol., 11, 2015
|
|
6TTV
 
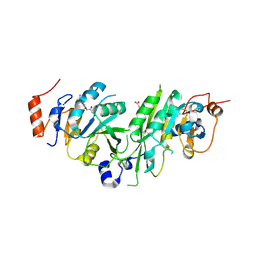 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 3 (ASI_M3M_138) | | 分子名称: | (2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolane-2-carboxamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-12-30 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
2JJV
 
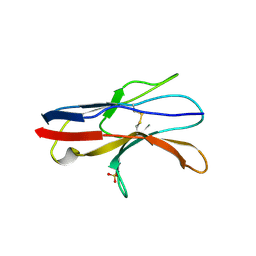 | | Structure of human signal regulatory protein (sirp) beta(2) | | 分子名称: | CHLORIDE ION, SIGNAL-REGULATORY PROTEIN BETA 1., SULFATE ION | | 著者 | Hatherley, D, Graham, S.C, Turner, J, Harlos, K, Stuart, D.I, Barclay, A.N. | | 登録日 | 2008-04-22 | | 公開日 | 2008-08-05 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Paired Receptor Specificity Explained by Structures of Signal Regulatory Proteins Alone and Complexed with Cd47.
Mol.Cell, 31, 2008
|
|
2O9T
 
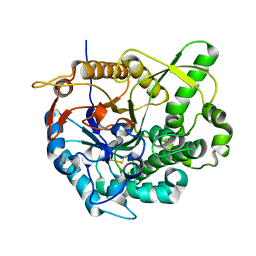 | |
4E5X
 
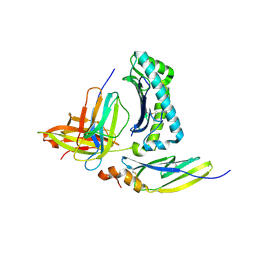 | |
4Z82
 
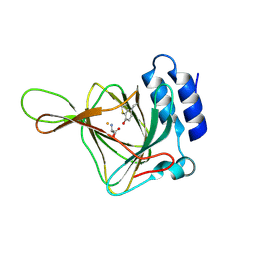 | | Cysteine bound rat cysteine dioxygenase C164S variant at pH 8.1 | | 分子名称: | CYSTEINE, Cysteine dioxygenase type 1, FE (II) ION | | 著者 | Fellner, M, Tchesnokov, E.P, Siakkou, E, Rutledge, M.T, Kanitz, M, Jameson, G.N.L, Wilbanks, S.M. | | 登録日 | 2015-04-08 | | 公開日 | 2016-06-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Influence of cysteine 164 on active site structure in rat cysteine dioxygenase.
J.Biol.Inorg.Chem., 21, 2016
|
|
4Z85
 
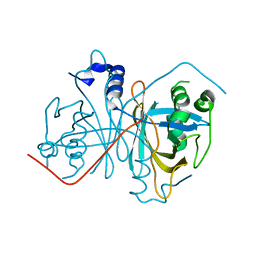 | |
4ZOA
 
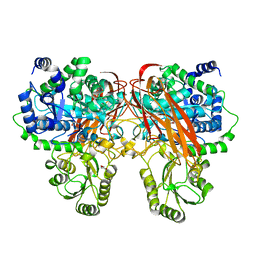 | | Crystal Structure of beta-glucosidase from Listeria innocua in complex with isofagomine | | 分子名称: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, DI(HYDROXYETHYL)ETHER, Lin1840 protein, ... | | 著者 | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | 登録日 | 2015-05-06 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
6T6C
 
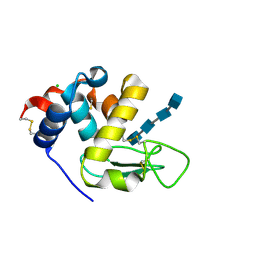 | | Complex with chitin oligomer of C-type lysozyme from the upper gastrointestinal tract of Opisthocomus hoatzin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C | | 著者 | Taylor, E.J, Skjot, M, Skov, L.K, Klausen, M, De Maria, L, Gippert, G.P, Turkenburg, J.P, Davies, G.J, Wilson, K.S. | | 登録日 | 2019-10-18 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | The C-Type Lysozyme from the upper Gastrointestinal Tract of Opisthocomus hoatzin, the Stinkbird.
Int J Mol Sci, 20, 2019
|
|
4E91
 
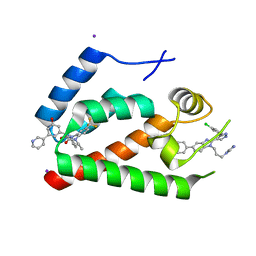 | | Crystal Structure of the N-Terminal Domain of HIV-1 Capsid in Complex With Inhibitor BD3 | | 分子名称: | (3S)-1-ethyl-3-[3-hydroxy-5-(pyridin-3-yl)phenyl]-5-phenyl-7-(trifluoromethyl)-1H-1,5-benzodiazepine-2,4(3H,5H)-dione, 4-{2-[5-(3-chlorophenyl)-1H-pyrazol-4-yl]-1-[3-(1H-imidazol-1-yl)propyl]-1H-benzimidazol-5-yl}benzoic acid, Gag protein, ... | | 著者 | Lemke, C.T. | | 登録日 | 2012-03-20 | | 公開日 | 2012-04-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Distinct Effects of Two HIV-1 Capsid Assembly Inhibitor Families That Bind the Same Site within the N-Terminal Domain of the Viral CA Protein.
J.Virol., 86, 2012
|
|
4Z9G
 
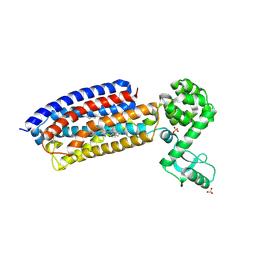 | | Crystal structure of human corticotropin-releasing factor receptor 1 (CRF1R) in complex with the antagonist CP-376395 in a hexagonal setting with translational non-crystallographic symmetry | | 分子名称: | 3,6-dimethyl-N-(pentan-3-yl)-2-(2,4,6-trimethylphenoxy)pyridin-4-amine, Corticotropin-releasing factor receptor 1,Lysozyme,Corticotropin-releasing factor receptor 1, OLEIC ACID, ... | | 著者 | Dore, A.S, Bortolato, A, Hollenstein, K, Cheng, R.K.Y, Read, R.J, Marshall, F.H. | | 登録日 | 2015-04-10 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.183 Å) | | 主引用文献 | Decoding Corticotropin-Releasing Factor Receptor Type 1 Crystal Structures.
Curr Mol Pharmacol, 10, 2017
|
|
