6HZR
 
 | | Apo structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 | | 分子名称: | Peptidoglycan D,D-transpeptidase FtsI | | 著者 | Bellini, D, Dowson, C.G. | | 登録日 | 2018-10-23 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.19 Å) | | 主引用文献 | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
6HZQ
 
 | | Apo structure of TP domain from Escherichia coli Penicillin-Binding Protein 3 | | 分子名称: | Peptidoglycan D,D-transpeptidase FtsI | | 著者 | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | 登録日 | 2018-10-23 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
6HZO
 
 | | Apo structure of TP domain from Haemophilus influenzae Penicillin-Binding Protein 3 | | 分子名称: | FtsI | | 著者 | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | 登録日 | 2018-10-23 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
6HZJ
 
 | | Apo structure of TP domain from clinical penicillin-resistant mutant Neisseria gonorrhoea strain 6140 Penicillin-Binding Protein 2 (PBP2) | | 分子名称: | Probable peptidoglycan D,D-transpeptidase PenA | | 著者 | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | 登録日 | 2018-10-23 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
6HZI
 
 | |
6HUH
 
 | | CRYSTAL STRUCTURE OF OXA-427 class D BETA-LACTAMASE | | 分子名称: | Beta-lactamase, SULFATE ION | | 著者 | Zavala, A, Retailleau, P, Bogaerts, P, Glupczynski, Y, Naas, T, Iorga, B. | | 登録日 | 2018-10-08 | | 公開日 | 2019-10-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF CMY-OXA-427-HisTag BETA-LACTAMASE
To be published
|
|
6HR9
 
 | |
6HR6
 
 | |
6HR4
 
 | |
6HOO
 
 | | Crystal Structure of Rationally Designed OXA-48loop18 beta-lactamase | | 分子名称: | Beta-lactamase,OXA-48loop18,Beta-lactamase, FLUORIDE ION, GLYCEROL, ... | | 著者 | Zavala, A, Retailleau, P, Dabos, L, Naas, T, Iorga, B. | | 登録日 | 2018-09-17 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Substrate Specificity of OXA-48 after beta 5-beta 6 Loop Replacement.
Acs Infect Dis., 6, 2020
|
|
6HB8
 
 | | Crystal structure of OXA-517 beta-lactamase | | 分子名称: | 1,2-ETHANEDIOL, 2-ETHOXYETHANOL, Beta-lactamase, ... | | 著者 | Raczynska, J.E, Dabos, L, Zavala, A, Retailleau, P, Iorga, B, Jaskolski, M, Naas, T. | | 登録日 | 2018-08-09 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Genetic, biochemical and structural characterization of OXA-517, an OXA-48-like extended-spectrum cephalosporins and carbapenems-hydrolyzing beta-lactamase
To Be Published
|
|
6H5O
 
 | |
6GOA
 
 | |
6G9S
 
 | | Structural basis for the inhibition of E. coli PBP2 | | 分子名称: | (3~{R},6~{S})-6-(aminomethyl)-4-(1,3-oxazol-5-yl)-3-(sulfooxyamino)-3,6-dihydro-2~{H}-pyridine-1-carboxylic acid, Peptidoglycan D,D-transpeptidase MrdA | | 著者 | Ruff, M, Levy, N. | | 登録日 | 2018-04-11 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
6G9P
 
 | |
6G9F
 
 | | Structural basis for the inhibition of E. coli PBP2 | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Peptidoglycan D,D-transpeptidase MrdA | | 著者 | Ruff, M, Levy, N. | | 登録日 | 2018-04-10 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
6G88
 
 | | Crystal structure of Enterococcus Faecium D63r Penicillin-Binding protein 5 (PBP5fm) | | 分子名称: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyrrolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Low affinity penicillin-binding protein 5 (PBP5), SULFATE ION | | 著者 | Sauvage, E, El Gachi, M, Herman, R, Kerff, F, Charlier, P. | | 登録日 | 2018-04-08 | | 公開日 | 2019-04-24 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural basis of inactivation of Enterococcus faecium penicillin binding protein 5 by ceftobiprole.
To Be Published
|
|
6G0K
 
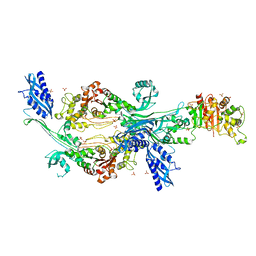 | | Crystal structure of Enterococcus faecium D63r Penicillin-Binding protein 5 (PBP5fm) | | 分子名称: | Low affinity penicillin-binding protein 5 (PBP5), SULFATE ION | | 著者 | Sauvage, E, El Gachi, M, Herman, R, Kerff, F, Charlier, P. | | 登録日 | 2018-03-19 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis of inactivation of Enterococcus faecium penicillin binding protein 5 by ceftobiprole.
To Be Published
|
|
6C7A
 
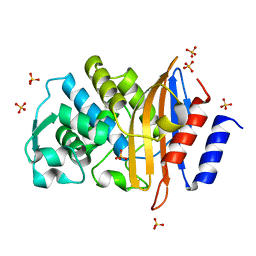 | | Conformational Changes in a Class A Beta lactamase that Prime it for Catalysis | | 分子名称: | Beta-lactamase Toho-1, SULFATE ION | | 著者 | Coates, L, Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M. | | 登録日 | 2018-01-22 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
6C79
 
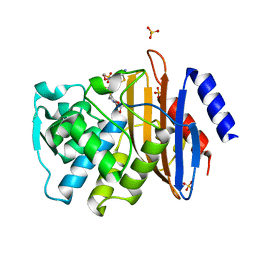 | | Conformational Changes in a Class A Beta lactamase that Prime it for Catalysis | | 分子名称: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase Toho-1, SULFATE ION | | 著者 | Coates, L, Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M. | | 登録日 | 2018-01-22 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
6C78
 
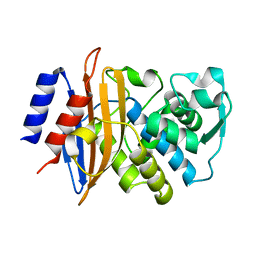 | | Substrate Binding Induces Conformational Changes In A Class A Beta Lactamase That Primes It For Catalysis | | 分子名称: | Beta-lactamase Toho-1 | | 著者 | Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M, Coates, L. | | 登録日 | 2018-01-22 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | NEUTRON DIFFRACTION (1.75 Å) | | 主引用文献 | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
6BSR
 
 | | Crystal structure of penicillin-binding protein 4 (PBP4) from Enterococcus faecalis in the benzylpenicillin bound form. | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | 登録日 | 2017-12-04 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6BSQ
 
 | | Enterococcus faecalis Penicillin Binding Protein 4 (PBP4) | | 分子名称: | CHLORIDE ION, GLYCEROL, PBP4 protein | | 著者 | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | 登録日 | 2017-12-04 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6B22
 
 | | Crystal structure OXA-24 beta-lactamase complexed with WCK 4234 by co-crystallization | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carbonitrile, Beta-lactamase, CHLORIDE ION | | 著者 | van den Akker, F, Nguyen, N.Q. | | 登録日 | 2017-09-19 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
5WIB
 
 | | Structure of Acinetobacter baumannii carbapenemase OXA-239 K82D bound to imipenem | | 分子名称: | Imipenem, OXA-239 | | 著者 | Harper, T.M, June, C.M, Powers, R.A, Leonard, D.A. | | 登録日 | 2017-07-19 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Multiple substitutions lead to increased loop flexibility and expanded specificity in Acinetobacter baumannii carbapenemase OXA-239.
Biochem. J., 475, 2018
|
|
