6DR0
 
 | | Class 5 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | 著者 | Hite, R.K, Paknejad, N. | | 登録日 | 2018-06-11 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (4.47 Å) | | 主引用文献 | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4QHO
 
 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with CCO10 | | 分子名称: | Alpha-ketoglutarate-dependent dioxygenase FTO, N-{[3-hydroxy-6-(naphthalen-1-yl)pyridin-2-yl]carbonyl}glycine, ZINC ION | | 著者 | Aik, W.S, Clunie-O'Connor, C, McDonough, M.A, Schofield, C.J. | | 登録日 | 2014-05-28 | | 公開日 | 2015-06-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structure-Based Design of Selective Fat Mass and Obesity Associated Protein (FTO) Inhibitors.
J.Med.Chem., 64, 2021
|
|
6VND
 
 | | Quaternary Complex of human dihydroorotate dehydrogenase (DHODH) with flavin mononucleotide (FMN), orotic acid and AG-636 | | 分子名称: | 1-methyl-5-(2'-methyl[1,1'-biphenyl]-4-yl)-1H-benzotriazole-7-carboxylic acid, ACETATE ION, CHLORIDE ION, ... | | 著者 | Padyana, A, Jin, L. | | 登録日 | 2020-01-29 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Selective Vulnerability to Pyrimidine Starvation in Hematologic Malignancies Revealed by AG-636, a Novel Clinical-Stage Inhibitor of Dihydroorotate Dehydrogenase.
Mol.Cancer Ther., 19, 2020
|
|
6DU7
 
 | |
7PG5
 
 | | Crystal Structure of PI3Kalpha | | 分子名称: | GLYCEROL, PHOSPHATE ION, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | 著者 | Gong, G, Pinotsis, N, Williams, R.L, Vanhaesebroeck, B. | | 登録日 | 2021-08-13 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.20029068 Å) | | 主引用文献 | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
6VON
 
 | | Chloroplast ATP synthase (R1, CF1FO) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | 著者 | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | 登録日 | 2020-01-30 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6VOL
 
 | | Chloroplast ATP synthase (R2, CF1FO) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | 著者 | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | 登録日 | 2020-01-30 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.06 Å) | | 主引用文献 | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
4K90
 
 | | Extracellular metalloproteinase from Aspergillus | | 分子名称: | BORIC ACID, CALCIUM ION, Extracellular metalloproteinase mep, ... | | 著者 | Fernandez, D, Russi, S, Vendrell, J, Monod, M, Pallares, I. | | 登録日 | 2013-04-19 | | 公開日 | 2013-10-23 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A functional and structural study of the major metalloprotease secreted by the pathogenic fungus Aspergillus fumigatus.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7SMP
 
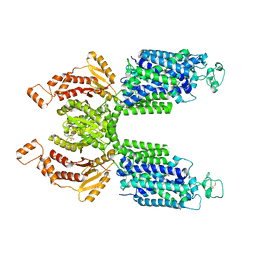 | | CryoEM structure of NKCC1 Bu-I | | 分子名称: | 3-(butylamino)-4-phenoxy-5-sulfamoylbenzoic acid, POTASSIUM ION, Solute carrier family 12 member 2 | | 著者 | Moseng, M.A. | | 登録日 | 2021-10-26 | | 公開日 | 2022-09-28 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
4KAO
 
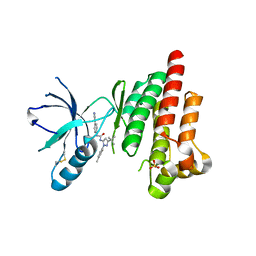 | | FOCAL ADHESION KINASE CATALYTIC DOMAIN IN COMPLEX WITH 1-(5-tert-Butyl-2-p-tolyl-2H-pyrazol-3-yl)-3-(4-pyridin-3- yl-phenyl)-urea | | 分子名称: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[4-(pyridin-3-yl)phenyl]urea, Focal adhesion kinase 1, SULFATE ION | | 著者 | Musil, D, Graedler, U, Heinrich, T, Lehmann, M, Dresing, V. | | 登録日 | 2013-04-22 | | 公開日 | 2013-09-11 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Fragment-based discovery of focal adhesion kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6DIH
 
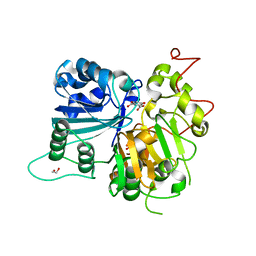 | | Crystal structure of Tdp1 catalytic domain in complex with Sigma Aldrich compound PH004941 | | 分子名称: | 1,2-ETHANEDIOL, 4-hydroxybenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | 著者 | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | 登録日 | 2018-05-23 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
7PG6
 
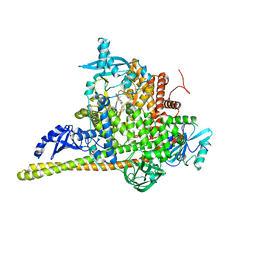 | | Crystal Structure of PI3Kalpha in complex with the inhibitor NVP-BYL719 | | 分子名称: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | 著者 | Gong, G, Pinotsis, N, Williams, R.L, Vanhaesebroeck, B. | | 登録日 | 2021-08-13 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.49943733 Å) | | 主引用文献 | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
6VVO
 
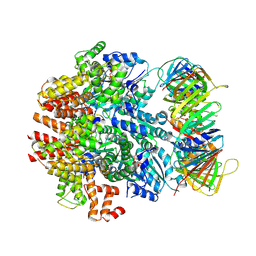 | | Structure of the human clamp loader (Replication Factor C, RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen, PCNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Gaubitz, C, Liu, X, Stone, N.P, Kelch, B.A. | | 登録日 | 2020-02-18 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of the human clamp loader bound to the sliding clamp: a further twist on AAA+ mechanism
Biorxiv, 2020
|
|
1ZAL
 
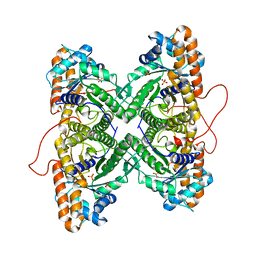 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle in complex with partially disordered tagatose-1,6-bisphosphate, a weak competitive inhibitor | | 分子名称: | Fructose-bisphosphate aldolase A, PHOSPHATE ION | | 著者 | St-Jean, M, Lafrance-Vanasse, J, Liotard, B, Sygusch, J. | | 登録日 | 2005-04-06 | | 公開日 | 2005-05-10 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | High Resolution Reaction Intermediates of Rabbit Muscle Fructose-1,6-bisphosphate Aldolase: substrate cleavage and induced fit.
J.Biol.Chem., 280, 2005
|
|
4QRI
 
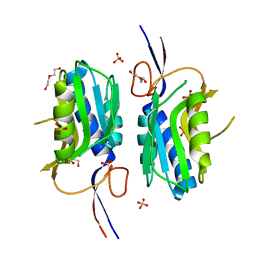 | | 2.35 Angstrom resolution crystal structure of hypoxanthine-guanine-xanthine phosphoribosyltransferase from Leptospira interrogans serovar Copenhageni str. Fiocruz L1-130 | | 分子名称: | DI(HYDROXYETHYL)ETHER, Hypoxanthine-guanine-xanthine phosphoribosyltransferase, SULFATE ION | | 著者 | Halavaty, A.S, Minasov, G, Dubrovska, I, Flores, K.J, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-07-01 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | 2.35 Angstrom resolution crystal structure of hypoxanthine-guanine-xanthine phosphoribosyltransferase from Leptospira interrogans serovar Copenhageni str. Fiocruz L1-130
To be Published
|
|
6DZF
 
 | |
6DZP
 
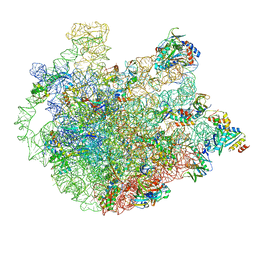 | | Cryo-EM Structure of Mycobacterium smegmatis C(minus) 50S ribosomal subunit | | 分子名称: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | 著者 | Sharma, M.R, Li, Y, Korripella, R, Yang, Y, Kaushal, P.S, Lin, Q, Wade, J.T, Gray, A.G, Derbyshire, K.M, Agrawal, R.K, Ojha, A. | | 登録日 | 2018-07-05 | | 公開日 | 2018-10-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | Zinc depletion induces ribosome hibernation in mycobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7JIC
 
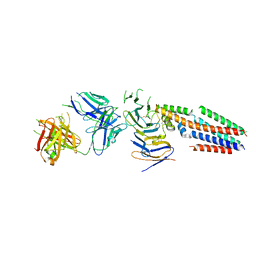 | | Structure of human CD19-CD81 co-receptor complex bound to coltuximab Fab fragment | | 分子名称: | B-lymphocyte antigen CD19, CD81 antigen, Coltuximab Heavy Chain, ... | | 著者 | Susa, K.J, Rawson, S, Kruse, A.C, Blacklow, S.C. | | 登録日 | 2020-07-23 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structure of the B cell co-receptor CD19 bound to the tetraspanin CD81
Science, 371, 2021
|
|
6DYS
 
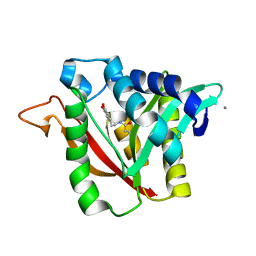 | | C-terminal condensation domain of Ebony in complex with beta-alanyl-dopamine | | 分子名称: | CALCIUM ION, Ebony, N-[2-(3,4-dihydroxyphenyl)ethyl]-beta-alaninamide | | 著者 | Izore, T, Tailhades, J, Hansen, M.H, Kaczmarski, J.A, Jackson, C.J, Cryle, M.J. | | 登録日 | 2018-07-02 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Drosophila melanogasternonribosomal peptide synthetase Ebony encodes an atypical condensation domain.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6W1V
 
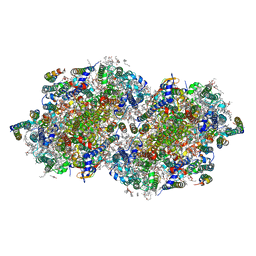 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.09 Angstrom resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | 登録日 | 2020-03-04 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6DCF
 
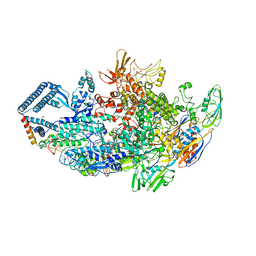 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase and bound to kanglemycin A | | 分子名称: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | 著者 | Lilic, M, Darst, S.A, Campbell, E.A. | | 登録日 | 2018-05-06 | | 公開日 | 2018-09-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.
Nat Commun, 9, 2018
|
|
6W2P
 
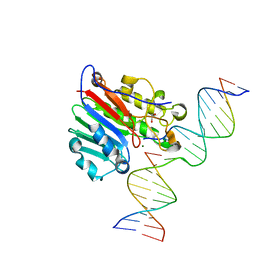 | | APE1 endonuclease product complex L104R | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | 著者 | Freudenthal, B.D, Whitaker, A.M. | | 登録日 | 2020-03-06 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
7JLW
 
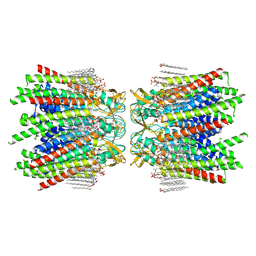 | | Sheep Connexin-50 at 2.5 angstroms resolution, Lipid Class 1 | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | 著者 | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | 登録日 | 2020-07-30 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
6W47
 
 | |
7SK4
 
 | | Cryo-EM structure of ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ, an intracellular Fab, and an extracellular Fab | | 分子名称: | Atypical chemokine receptor 3, CHOLESTEROL, CID24 Fab heavy chain, ... | | 著者 | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | 登録日 | 2021-10-19 | | 公開日 | 2022-07-27 | | 最終更新日 | 2025-06-04 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
