7XUG
 
 | |
7XUI
 
 | |
217D
 
 | |
1L45
 
 | |
1GFG
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-12-04 | | 公開日 | 2000-12-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFR
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-12-04 | | 公開日 | 2000-12-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1L55
 
 | |
1LGV
 
 | | Structure of a Human Bence-Jones Dimer Crystallized in U.S. Space Shuttle Mission STS-95: 100K | | 分子名称: | IMMUNOGLOBULIN LAMBDA LIGHT CHAIN | | 著者 | Terzyan, S.S, DeWitt, C.R, Ramsland, P.A, Bourne, P.C, Edmundson, A.B. | | 登録日 | 2002-04-16 | | 公開日 | 2003-07-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Comparison of the three-dimensional structures of a human Bence-Jones dimer
crystallized on Earth and aboard US Space Shuttle Mission STS-95
J.MOL.RECOG., 16, 2003
|
|
5CLE
 
 | |
4PTG
 
 | | Structure of a carboxamine compound (26) (2-{2-[(CYCLOPROPYLCARBONYL)AMINO]PYRIDIN-4-YL}-4-METHOXYPYRIMIDINE-5-CARBOXAMIDE) to GSK3b | | 分子名称: | 2-{2-[(cyclopropylcarbonyl)amino]pyridin-4-yl}-4-methoxypyrimidine-5-carboxamide, Glycogen synthase kinase-3 beta | | 著者 | Lewis, H.A, Sivaprakasam, P, Kish, K, Pokross, M, Dubowchik, G.M. | | 登録日 | 2014-03-10 | | 公開日 | 2015-04-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.361 Å) | | 主引用文献 | Discovery of new acylaminopyridines as GSK-3 inhibitors by a structure guided in-depth exploration of chemical space around a pyrrolopyridinone core.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5CLV
 
 | | Crystal Structure of KorA-operator DNA complex (KorA-OA) | | 分子名称: | 5'-D(CP*CP*AP*AP*GP*TP*TP*TP*AP*GP*CP*TP*AP*AP*AP*CP*TP*TP*GP*GP*)-3', TrfB transcriptional repressor protein | | 著者 | White, S.A, Hyde, E.I, Rajasekar, K.V. | | 登録日 | 2015-07-16 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
6KHD
 
 | | Crystal structure of CLK1 in complex with CX-4945 | | 分子名称: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK1 | | 著者 | Lee, J.Y, Yun, J.S, Jin, H, Chang, J.H. | | 登録日 | 2019-07-15 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945.
Biomed Res Int, 2019, 2019
|
|
2CBL
 
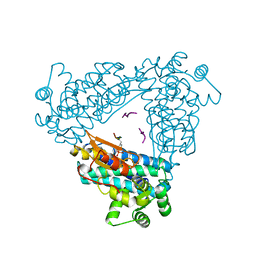 | | N-TERMINAL DOMAIN OF CBL IN COMPLEX WITH ITS BINDING SITE ON ZAP-70 | | 分子名称: | CALCIUM ION, PROTO-ONCOGENE CBL, ZAP-70 | | 著者 | Meng, W, Sawasdikosol, S, Burakoff, S.J, Eck, M.J. | | 登録日 | 1998-08-28 | | 公開日 | 1999-05-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase.
Nature, 398, 1999
|
|
1L69
 
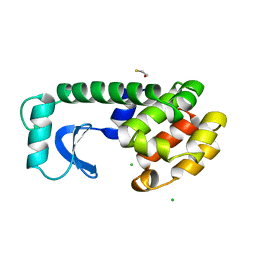 | |
1GN9
 
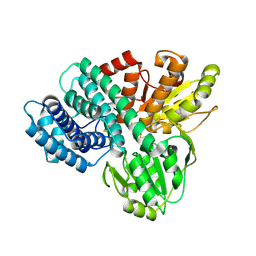 | | Hybrid Cluster Protein from Desulfovibrio desulfuricans ATCC 27774 X-ray structure at 2.6A resolution using synchrotron radiation at a wavelength of 1.722A | | 分子名称: | HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | 著者 | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | 登録日 | 2001-10-04 | | 公開日 | 2002-04-11 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Hybrid Cluster Proteins (Hcps) from Desulfovibrio Desulfuricans Atcc 27774 and Desulfovibrio Vulgaris (Hildenborough): X-Ray Structures at 1.25 A Resolution Using Synchrotron Radiation.
J.Biol.Inorg.Chem., 7, 2002
|
|
5CL3
 
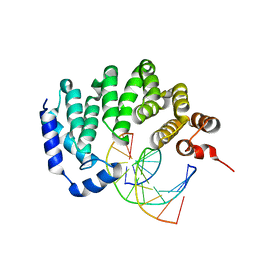 | |
5CL8
 
 | |
2C7I
 
 | |
1ZUI
 
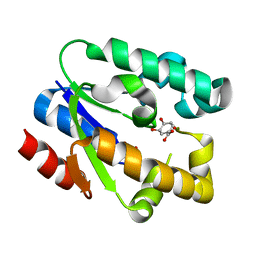 | | Structural Basis for Shikimate-binding Specificity of Helicobacter pylori Shikimate Kinase | | 分子名称: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, PHOSPHATE ION, Shikimate kinase | | 著者 | Cheng, W.C, Chang, Y.N, Wang, W.C. | | 登録日 | 2005-05-31 | | 公開日 | 2006-05-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for shikimate-binding specificity of Helicobacter pylori shikimate kinase
J.Bacteriol., 187, 2005
|
|
2CLQ
 
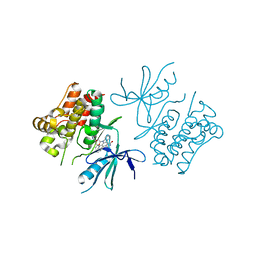 | | Structure of mitogen-activated protein kinase kinase kinase 5 | | 分子名称: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, STAUROSPORINE | | 著者 | Bunkoczi, G, Salah, E, Fedorov, O, Pike, A, Gileadi, O, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Knapp, S. | | 登録日 | 2006-04-28 | | 公開日 | 2006-05-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Functional Characterization of the Human Protein Kinase Ask1.
Structure, 15, 2007
|
|
1L85
 
 | |
1L8H
 
 | | DNA PROTECTION AND BINDING BY E. COLI DPS PROTEIN | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA PROTECTION DURING STARVATION PROTEIN, POTASSIUM ION | | 著者 | Luo, J, Liu, D, White, M.A, Fox, R.O. | | 登録日 | 2002-03-20 | | 公開日 | 2003-06-24 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | DNA Protection and Binding by E. Coli Dps Protein
To be Published
|
|
1FWC
 
 | |
1KY1
 
 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | 著者 | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | 登録日 | 2002-02-01 | | 公開日 | 2003-06-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
5CRA
 
 | | Structure of the SdeA DUB Domain | | 分子名称: | METHYL 4-AMINOBUTANOATE, Polyubiquitin-B, SULFATE ION, ... | | 著者 | Sheedlo, M.J, Qiu, J, Luo, Z.Q, Das, C. | | 登録日 | 2015-07-22 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
