5F5A
 
 | | Crystal Structure of human JMJD2D complexed with KDOAM16 | | 分子名称: | 1,2-ETHANEDIOL, 2-[(furan-2-ylmethylamino)methyl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4D, ... | | 著者 | Krojer, T, Vollmar, M, Crawley, L, Bradley, A.R, Szykowska, A, Ruda, G.F, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | 登録日 | 2015-12-04 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F2W
 
 | | Crystal structure of human KDM4A in complex with compound 16 | | 分子名称: | 1,2-ETHANEDIOL, 2-(2-azanyl-1,3-thiazol-4-yl)pyridine-4-carboxamide, DIMETHYL SULFOXIDE, ... | | 著者 | Le Bihan, Y.-V, Dempster, S, Westwood, I.M, van Montfort, R.L.M. | | 登録日 | 2015-12-02 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
4LWB
 
 | | Structure of Bacillus subtilis nitric oxide synthase in complex with 6-((((3R,5S)-5-(((6-amino-4-methylpyridin-2-yl)methoxy)methyl)pyrrolidin-3-yl)oxy)methyl)-4-methylpyridin-2-amine | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-({[(3R,5S)-5-{[(6-amino-4-methylpyridin-2-yl)methoxy]methyl}pyrrolidin-3-yl]oxy}methyl)-4-methylpyridin-2-amine, CHLORIDE ION, ... | | 著者 | Holden, J.K, Li, H, Poulos, T.L. | | 登録日 | 2013-07-26 | | 公開日 | 2013-10-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural and biological studies on bacterial nitric oxide synthase inhibitors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7Q1S
 
 | | A de novo designed hetero-dimeric antiparallel coiled coil apCC-Di-AB_var | | 分子名称: | 1,2-ETHANEDIOL, apCC-Di-A_var, apCC-Di-B_var | | 著者 | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | 登録日 | 2021-10-20 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
7PHM
 
 | |
3F36
 
 | | Apoferritin: complex with 2-isopropylphenol | | 分子名称: | 2-(1-methylethyl)phenol, CADMIUM ION, Ferritin light chain, ... | | 著者 | Vedula, L.S, Economou, N.J, Rossi, M.J, Eckenhoff, R.G, Loll, P.J. | | 登録日 | 2008-10-30 | | 公開日 | 2009-07-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A unitary anesthetic binding site at high resolution.
J.Biol.Chem., 284, 2009
|
|
3A2V
 
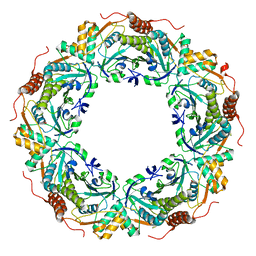 | | Peroxiredoxin (C207S) from Aeropyrum pernix K1 complexed with hydrogen peroxide | | 分子名称: | PEROXIDE ION, Probable peroxiredoxin | | 著者 | Nakamura, T, Kado, Y, Yamaguchi, F, Ishikawa, K, Matsumura, H, Inoue, T. | | 登録日 | 2009-06-04 | | 公開日 | 2009-10-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
7K1D
 
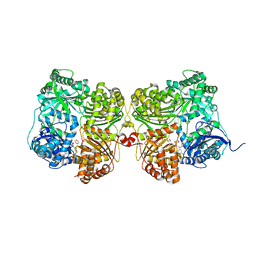 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM_77291 | | 分子名称: | (3R)-3-{4-[(3R)-4-(3,4-difluorobenzene-1-carbonyl)morpholin-3-yl]-1H-1,2,3-triazol-1-yl}-N-hydroxy-4-(naphthalen-2-yl)butanamide, 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Liang, W.G, Deprez, R, Bosc, D, Tang, W. | | 登録日 | 2020-09-07 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 2
To Be Published
|
|
2RV9
 
 | |
4LO8
 
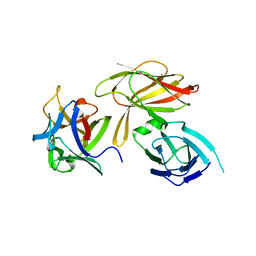 | | HA70(D3)-HA17 | | 分子名称: | HA-17, HA-70 | | 著者 | Lee, K, Gu, S, Jin, L, Le, T.T, Cheng, L.W, Strotmeier, J, Kruel, A.M, Yao, G, Perry, K, Rummel, A, Jin, R. | | 登録日 | 2013-07-12 | | 公開日 | 2013-10-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity.
Plos Pathog., 9, 2013
|
|
3F3P
 
 | | Crystal structure of the nucleoporin pair Nup85-Seh1, space group P21212 | | 分子名称: | Nucleoporin NUP85, Nucleoporin SEH1 | | 著者 | Debler, E.W, Hseo, H, Ma, Y, Blobel, G, Hoelz, A. | | 登録日 | 2008-10-31 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | A fence-like coat for the nuclear pore membrane.
Mol.Cell, 32, 2008
|
|
5C09
 
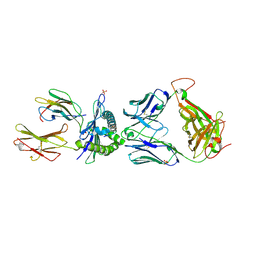 | | HLA class I histocompatibility antigen | | 分子名称: | 1E6 TCR Alpha Chain, 1E6 TCR Beta Chain, Beta-2-microglobulin, ... | | 著者 | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | 登録日 | 2015-06-12 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.475 Å) | | 主引用文献 | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J.Clin.Invest., 126, 2016
|
|
7K1F
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM_88558 | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, 3,4-difluoro-N-({1-[(2R)-4-(hydroxyamino)-4-oxo-1-(quinolin-7-yl)butan-2-yl]-1H-1,2,3-triazol-4-yl}methyl)benzamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Liang, W.G, Deprez, R, Bosc, D, Tang, W. | | 登録日 | 2020-09-07 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 4
To Be Published
|
|
7Q1T
 
 | | A de novo designed hetero-dimeric antiparallel coiled coil apCC-Di-AB | | 分子名称: | N-PROPANOL, SULFATE ION, apCC-Di-A, ... | | 著者 | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | 登録日 | 2021-10-20 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
7K1E
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM_88646 | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, 3,4-difluoro-N-[(1S)-1-{1-[(2R)-4-(hydroxyamino)-4-oxo-1-(5,6,7,8-tetrahydronaphthalen-2-yl)butan-2-yl]-1H-1,2,3-triazol-4-yl}ethyl]benzamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Liang, W.G, Deprez, R, Bosc, D, Tang, W. | | 登録日 | 2020-09-07 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 3
To Be Published
|
|
4LQJ
 
 | | Two minutes iron loaded frog M ferritin | | 分子名称: | CHLORIDE ION, FE (II) ION, Ferritin, ... | | 著者 | Mangani, S, Di Pisa, F, Pozzi, C, Turano, P, Lalli, D. | | 登録日 | 2013-07-18 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Time-lapse anomalous X-ray diffraction shows how Fe(2+) substrate ions move through ferritin protein nanocages to oxidoreductase sites.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7Q1Q
 
 | | De novo designed homo-dimeric antiparallel helices Homomer-S | | 分子名称: | ACETATE ION, Homomer-S | | 著者 | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | 登録日 | 2021-10-20 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
5C37
 
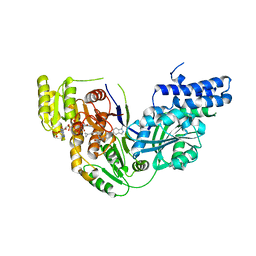 | | Structure of the beta-ketoacyl reductase domain of human fatty acid synthase bound to a spiro-imidazolone inhibitor | | 分子名称: | 6-{[(3R)-1-(cyclopropylcarbonyl)pyrrolidin-3-yl]methyl}-5-[4-(1-methyl-1H-indazol-5-yl)phenyl]-4,6-diazaspiro[2.4]hept-4-en-7-one, CHLORIDE ION, Fatty acid synthase, ... | | 著者 | Schubert, C, Milligan, C.M, Vo, K, Grasberger, B. | | 登録日 | 2015-06-17 | | 公開日 | 2016-06-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Design and synthesis of a series of bioavailable fatty acid synthase (FASN) KR domain inhibitors for cancer therapy.
Bioorg.Med.Chem.Lett., 28, 2018
|
|
5BYV
 
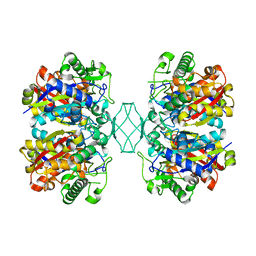 | | Crystal structure of MSM-13, a putative T1-like thiolase from Mycobacterium smegmatis | | 分子名称: | Beta-ketothiolase | | 著者 | Janardan, N, Harijan, R.K, Keima, T.R, Wierenga, R, Murthy, M.R.N. | | 登録日 | 2015-06-11 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.162 Å) | | 主引用文献 | Structural characterization of a mitochondrial 3-ketoacyl-CoA (T1)-like thiolase from Mycobacterium smegmatis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3FBM
 
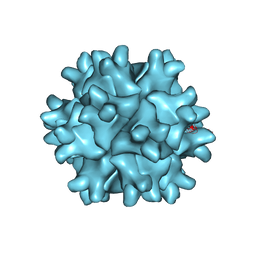 | | D431N Mutant VP2 Protein of Infectious Bursal Disease Virus; Derived T=1 Particles | | 分子名称: | CALCIUM ION, Polyprotein | | 著者 | Irigoyen, N, Garriga, D, Navarro, A, Verdaguer, N, Rodriguez, J.F, Caston, J.R. | | 登録日 | 2008-11-19 | | 公開日 | 2009-01-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Autoproteolytic Activity Derived from the Infectious Bursal Disease Virus Capsid Protein
J.Biol.Chem., 284, 2009
|
|
5C08
 
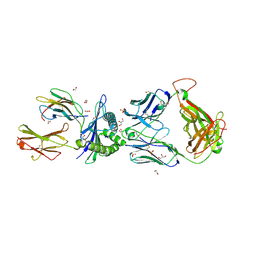 | | 1E6 TCR in Complex with HLA-A0e carrying RQWGPDPAAV | | 分子名称: | 1,2-ETHANEDIOL, 1E6 TCR Alpha Chain, 1E6 TCR Beta Chain, ... | | 著者 | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | 登録日 | 2015-06-12 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.332 Å) | | 主引用文献 | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J.Clin.Invest., 126, 2016
|
|
7JTA
 
 | | Crystal structure of a putative nuclease with anti-Cas9 activity from an uncultured Clostridia bacterium | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, NITRATE ION, NTF2-like nuclease/anti-CRISPR | | 著者 | Werther, R, Forsberg, K.J, Stoddard, B.L. | | 登録日 | 2020-08-17 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | The novel anti-CRISPR AcrIIA22 relieves DNA torsion in target plasmids and impairs SpyCas9 activity.
Plos Biol., 19, 2021
|
|
7Q10
 
 | |
5F39
 
 | | Crystal structure of human KDM4A in complex with compound 37 | | 分子名称: | 8-(1,3-thiazol-4-yl)-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, Lysine-specific demethylase 4A, ... | | 著者 | Le Bihan, Y.-V, Dempster, S, Westwood, I.M, van Montfort, R.L.M. | | 登録日 | 2015-12-02 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
3FC3
 
 | | Crystal structure of the beta-beta-alpha-Me type II restriction endonuclease Hpy99I | | 分子名称: | 5'-(*DCP*DTP*DCP*DGP*DAP*DCP*DGP*DTP*DAP*DGP*DA)-3', 5'-(*DTP*DAP*DCP*DGP*DTP*DCP*DGP*DAP*DGP*DTP*DC)-3', Restriction endonuclease Hpy99I, ... | | 著者 | Sokolowska, M, Czapinska, H, Bochtler, M. | | 登録日 | 2008-11-21 | | 公開日 | 2009-03-31 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of the beta beta alpha-Me type II restriction endonuclease Hpy99I with target DNA.
Nucleic Acids Res., 37, 2009
|
|
