3RHP
 
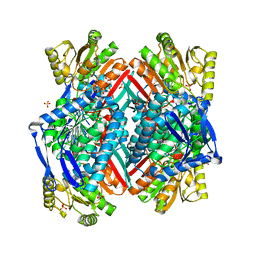 | |
3RI9
 
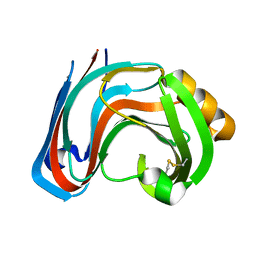 | | Xylanase C from Aspergillus kawachii F131W mutant | | 分子名称: | Endo-1,4-beta-xylanase 3 | | 著者 | Fushinobu, S, Uno, T, Kitaoka, M, Hayashi, K, Matsuzawa, H, Wakagi, T. | | 登録日 | 2011-04-13 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mutational analysis of fungal family 11 xylanases on pH optimum determination
J.APPL.GLYOSCI., 58, 2011
|
|
3R31
 
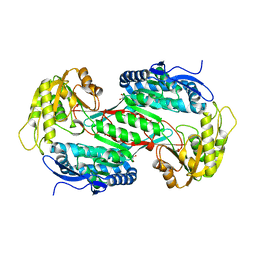 | |
3BEN
 
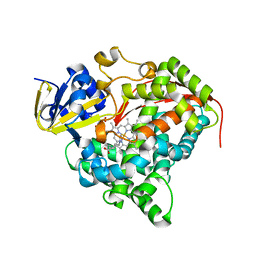 | |
1MAB
 
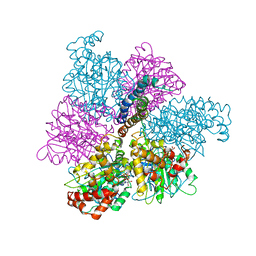 | | RAT LIVER F1-ATPASE | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Bianchet, M.A, Amzel, L.M. | | 登録日 | 1998-08-06 | | 公開日 | 1998-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The 2.8-A structure of rat liver F1-ATPase: configuration of a critical intermediate in ATP synthesis/hydrolysis.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6BN0
 
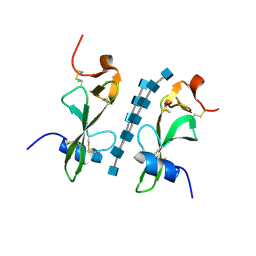 | | Avirulence protein 4 (Avr4) from Cladosporium fulvum bound to the hexasaccharide of chitin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Race-specific elicitor A4 | | 著者 | Hurlburt, N.K, Fisher, A.J. | | 登録日 | 2017-11-15 | | 公開日 | 2018-08-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure of the Cladosporium fulvum Avr4 effector in complex with (GlcNAc)6 reveals the ligand-binding mechanism and uncouples its intrinsic function from recognition by the Cf-4 resistance protein.
PLoS Pathog., 14, 2018
|
|
3R7M
 
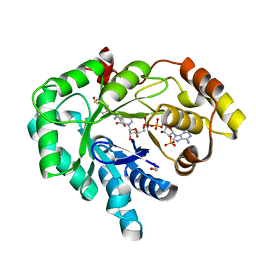 | | AKR1C3 complex with sulindac | | 分子名称: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Yosaatmadja, Y, Teague, R.M, Flanagan, J.U, Squire, C.J. | | 登録日 | 2011-03-22 | | 公開日 | 2012-05-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of three classes of non-steroidal anti-inflammatory drugs in complex with aldo-keto reductase 1C3.
Plos One, 7, 2012
|
|
3BA1
 
 | | Structure of hydroxyphenylpyruvate reductase from coleus blumei | | 分子名称: | Hydroxyphenylpyruvate reductase | | 著者 | Janiak, V, Klebe, G, Petersen, M, Heine, A. | | 登録日 | 2007-11-07 | | 公開日 | 2008-11-11 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Structure and substrate docking of a hydroxy(phenyl)pyruvate reductase from the higher plant Coleus blumei Benth.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3RQS
 
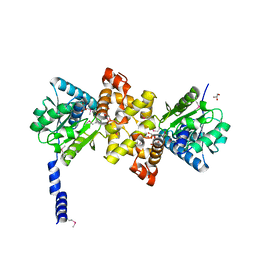 | | Crystal Structure of human L-3- Hydroxyacyl-CoA dehydrogenase (EC1.1.1.35) from mitochondria at the resolution 2.0 A, Northeast Structural Genomics Consortium Target HR487, Mitochondrial Protein Partnership | | 分子名称: | GLYCEROL, Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | | 著者 | Kuzin, A, Su, M, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | 登録日 | 2011-04-28 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target HR487
To be published
|
|
6RNE
 
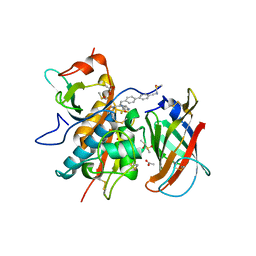 | | DPP1 in complex with inhibitor | | 分子名称: | (2~{S},4~{S})-~{N}-[(2~{S})-1-azanyl-3-[4-(4-cyanophenyl)phenyl]propan-2-yl]-4-fluoranyl-pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kack, H. | | 登録日 | 2019-05-08 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | DPP1 Inhibitors: Exploring the Role of Water in the S2 Pocket of DPP1 with Substituted Pyrrolidines.
Acs Med.Chem.Lett., 10, 2019
|
|
3RHD
 
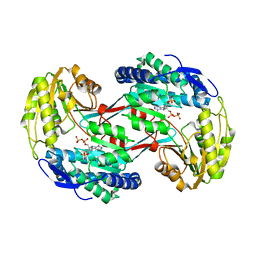 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661 complexed with NADP | | 分子名称: | Lactaldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-04-11 | | 公開日 | 2011-05-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661 complexed with NADP
To be Published
|
|
3RIL
 
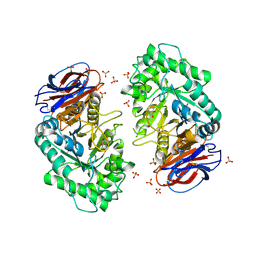 | | The acid beta-glucosidase active site exhibits plasticity in binding 3,4,5,6-tetrahydroxyazepane-based inhibitors: implications for pharmacological chaperone design for gaucher disease | | 分子名称: | (3S,4R,5R,6S)-azepane-3,4,5,6-tetrol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, ... | | 著者 | Orwig, S.D, Lieberman, R.L. | | 登録日 | 2011-04-13 | | 公開日 | 2012-03-14 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Binding of 3,4,5,6-tetrahydroxyazepanes to the acid-beta-glucosidase active site: implications for pharmacological chaperone design for Gaucher disease
Biochemistry, 50, 2011
|
|
3BAI
 
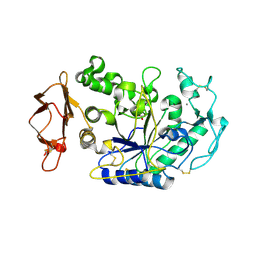 | | Human Pancreatic Alpha Amylase with Bound Nitrate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NITRATE ION, ... | | 著者 | Fredriksen, J.R, Maurus, R, Brayer, G.D. | | 登録日 | 2007-11-07 | | 公開日 | 2008-03-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
3BH4
 
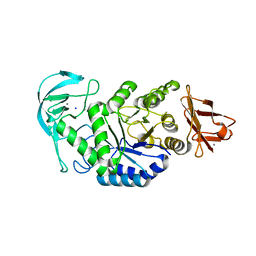 | | High resolution crystal structure of Bacillus amyloliquefaciens alpha-amylase | | 分子名称: | Alpha-amylase, CALCIUM ION, SODIUM ION | | 著者 | Alikhajeh, J, Khajeh, K, Ranjbar, B, Naderi-Manesh, H, Lin, Y.H, Liu, M.Y, Chen, C.J. | | 登録日 | 2007-11-28 | | 公開日 | 2008-12-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure of Bacillus amyloliquefaciens alpha-amylase at high resolution: implications for thermal stability.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3R1Z
 
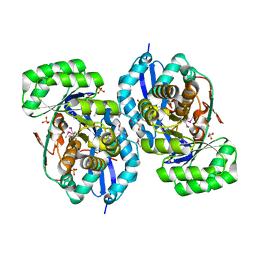 | | Crystal structure of NYSGRC enolase target 200555, a putative dipeptide epimerase from Francisella philomiragia : Complex with L-Ala-L-Glu and L-Ala-D-Glu | | 分子名称: | ALANINE, D-GLUTAMIC ACID, Enzyme of enolase superfamily, ... | | 著者 | Vetting, M.W, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-03-11 | | 公開日 | 2011-04-20 | | 最終更新日 | 2012-03-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6S0A
 
 | | Crystal Structure of Properdin (TSR domains N12 & 456) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Properdin, alpha-D-mannopyranose, ... | | 著者 | van den Bos, R.M, Pearce, N.M, Gros, P. | | 登録日 | 2019-06-14 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Insights Into Enhanced Complement Activation by Structures of Properdin and Its Complex With the C-Terminal Domain of C3b.
Front Immunol, 10, 2019
|
|
3ROS
 
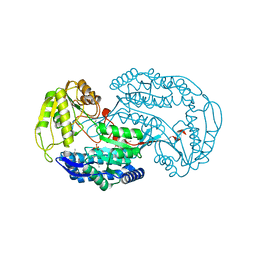 | |
6CX2
 
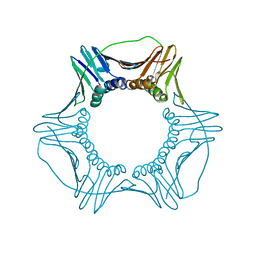 | | S177G Mutant of Yeast PCNA | | 分子名称: | Proliferating cell nuclear antigen | | 著者 | Powers, K.T. | | 登録日 | 2018-04-02 | | 公開日 | 2018-04-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.101 Å) | | 主引用文献 | Identification of New Mutations at the PCNA Subunit Interface that Block Translesion Synthesis.
PLoS ONE, 11, 2016
|
|
6RYF
 
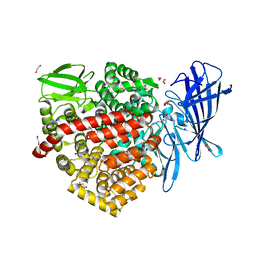 | | High-resolution crystal structure of ERAP1 in complex with 15mer phosphinic peptide | | 分子名称: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Giastas, P, Stratikos, E. | | 登録日 | 2019-06-10 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Mechanism for antigenic peptide selection by endoplasmic reticulum aminopeptidase 1.
Proc.Natl.Acad.Sci.USA, 2019
|
|
3C81
 
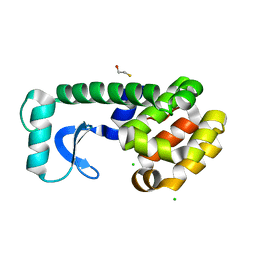 | |
3BNB
 
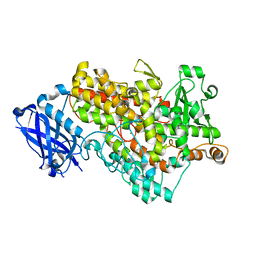 | | Lipoxygenase-1 (Soybean) I553L Mutant | | 分子名称: | FE (III) ION, Seed lipoxygenase-1 | | 著者 | Tomchick, D.R. | | 登録日 | 2007-12-14 | | 公開日 | 2008-04-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Enzyme structure and dynamics affect hydrogen tunneling: the impact of a remote side chain (I553) in soybean lipoxygenase-1.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C8S
 
 | |
3R3J
 
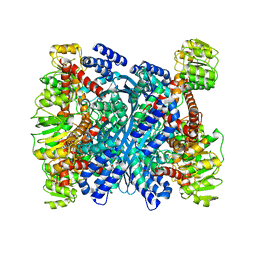 | | Kinetic and structural characterization of Plasmodium falciparum glutamate dehydrogenase 2 | | 分子名称: | Glutamate dehydrogenase | | 著者 | Zocher, K, Fritz-Wolf, K, Kehr, S, Rahlfs, S, Becker, K. | | 登録日 | 2011-03-16 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Biochemical and structural characterization of Plasmodium falciparum glutamate dehydrogenase 2.
Mol.Biochem.Parasitol., 183, 2012
|
|
6NIR
 
 | |
3BSG
 
 | |
