5IIB
 
 | | Crystal structure of red abalone egg VERL repeat 3 in complex with sperm lysin at 1.64 A resolution (crystal form II) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Egg-lysin, ... | | 著者 | Raj, I, Sadat Al-Hosseini, H, Nishimura, K, De Sanctis, D, Jovine, L. | | 登録日 | 2016-03-01 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5KWB
 
 | | Crystal Structure of the Receptor Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1 (HKU1 1A-CTD, 1.9 angstrom, molecular replacement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Spike glycoprotein, ... | | 著者 | Guan, H, Wojdyla, J.A, Wang, M, Cui, S. | | 登録日 | 2016-07-17 | | 公開日 | 2017-06-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1
Nat Commun, 8, 2017
|
|
5L6V
 
 | | Crystal structure of E. coli ADP-glucose pyrophosphorylase (AGPase) in complex with a negative allosteric regulator adenosine monophosphate (AMP) - AGPase*AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Glucose-1-phosphate adenylyltransferase, PHOSPHATE ION, ... | | 著者 | Cifuente, J.O, Albesa-Jove, D, Comino, N, Madariaga-Marcos, J, Agirre, J, Lopez-Fernandez, S, Garcia-Alija, M, Guerin, M.E. | | 登録日 | 2016-05-31 | | 公開日 | 2016-09-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.667 Å) | | 主引用文献 | Structural Basis of Glycogen Biosynthesis Regulation in Bacteria.
Structure, 24, 2016
|
|
5N6N
 
 | | CRYSTAL STRUCTURE OF THE 14-3-3:NEUTRAL TREHALASE NTH1 COMPLEX | | 分子名称: | CALCIUM ION, Neutral trehalase, Protein BMH1, ... | | 著者 | Alblova, M, Smidova, A, Obsilova, V, Obsil, T. | | 登録日 | 2017-02-15 | | 公開日 | 2017-11-01 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Molecular basis of the 14-3-3 protein-dependent activation of yeast neutral trehalase Nth1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5N6V
 
 | | Crystal structure of Neisseria polysaccharea amylosucrase mutant derived from Neutral genetic Drift-based engineering | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Amylosucrase, ... | | 著者 | Daude, D, Verges, A, Tranier, S. | | 登録日 | 2017-02-16 | | 公開日 | 2018-03-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Neutral Genetic Drift-Based Engineering of a Sucrose-Utilizing Enzyme toward Glycodiversification.
Acs Catalysis, 2019
|
|
5N7J
 
 | |
5O8L
 
 | | Crystal structure of Leuconostoc citreum NRRL B-1299 N-terminally truncated dextransucrase DSR-M in complex with sucrose | | 分子名称: | Alternansucrase, CALCIUM ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | 著者 | Claverie, M, Cioci, G, Remaud-simeon, M, Moulis, C, Tranier, S. | | 登録日 | 2017-06-13 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Investigations on the Determinants Responsible for Low Molar Mass Dextran Formation by DSR-M Dextransucrase
Acs Catalysis, 7, 2017
|
|
5TOY
 
 | |
5TXL
 
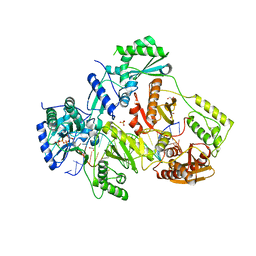 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DATP | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG) P*CP*GP*CP*CP*GP)-3'), ... | | 著者 | Das, K, Martinez, S.M, Arnold, E. | | 登録日 | 2016-11-17 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
5TXM
 
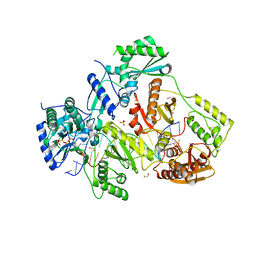 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DDATP | | 分子名称: | 1,2-ETHANEDIOL, 2',3'-dideoxyadenosine triphosphate, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | 著者 | Das, K, Martinez, S.M, Arnold, E. | | 登録日 | 2016-11-17 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
5TXN
 
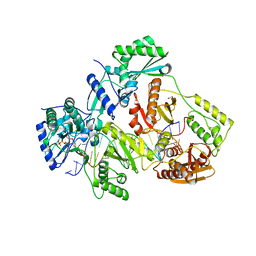 | | STRUCTURE OF Q151M MUTANT HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DATP | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | 著者 | Das, K, Martinez, S.M, Arnold, E. | | 登録日 | 2016-11-17 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
5TXO
 
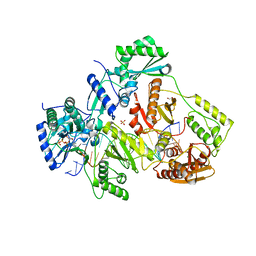 | | STRUCTURE OF Q151M complex (A62V, V75I, F77L, F116Y, Q151M) mutant HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DATP | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | 著者 | Das, K, Martinez, S.M, Arnold, E. | | 登録日 | 2016-11-17 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.546 Å) | | 主引用文献 | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
5TXP
 
 | | STRUCTURE OF Q151M complex (A62V, V75I, F77L, F116Y, Q151M) mutant HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DDATP | | 分子名称: | 1,2-ETHANEDIOL, 2',3'-dideoxyadenosine triphosphate, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | 著者 | Das, K, Martinez, S.M, Arnold, E. | | 登録日 | 2016-11-17 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
5UI2
 
 | | CRYSTAL STRUCTURE OF ORANGE CAROTENOID PROTEIN | | 分子名称: | (3'R)-3'-hydroxy-beta,beta-caroten-4-one, CHLORIDE ION, Orange carotenoid-binding protein, ... | | 著者 | KERFELD, C.A, SAWAYA, M.R, VISHNU, B, KROGMANN, D, YEATES, T.O. | | 登録日 | 2017-01-12 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of a cyanobacterial water-soluble carotenoid binding protein.
Structure, 11, 2003
|
|
5W75
 
 | |
5XN0
 
 | | HIV-1 reverse transcriptase Q151M:DNA binary complex | | 分子名称: | 38-MER DNA aptamer, GLYCEROL, Pol protein, ... | | 著者 | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | 登録日 | 2017-05-17 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.596 Å) | | 主引用文献 | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
5XN1
 
 | | HIV-1 reverse transcriptase Q151M:DNA:entecavir-triphosphate ternary complex | | 分子名称: | 38-MER DNA aptamer, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | 登録日 | 2017-05-17 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.446 Å) | | 主引用文献 | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
5XN2
 
 | | HIV-1 reverse transcriptase Q151M:DNA:dGTP ternary complex | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 38-MER DNA aptamer, GLYCEROL, ... | | 著者 | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | 登録日 | 2017-05-17 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.381 Å) | | 主引用文献 | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
5Z74
 
 | | Crystal structure of alkaline/neutral invertase InvB from Anabaena sp. PCC 7120 complexed with sucrose | | 分子名称: | Alr0819 protein, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | 著者 | Xie, J, Hu, H.X, Cai, K, Yang, F, Jiang, Y.L, Chen, Y, Zhou, C.Z. | | 登録日 | 2018-01-27 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural and enzymatic analyses of Anabaena heterocyst-specific alkaline invertase InvB.
FEBS Lett., 592, 2018
|
|
6BHJ
 
 | | Structure of HIV-1 Reverse Transcriptase Bound to a 38-mer Hairpin Template-Primer RNA-DNA Aptamer | | 分子名称: | 38-MER RNA-DNA Aptamer, GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | 著者 | Ruiz, F.X, Miller, M.T, Tuske, S, Das, K, Arnold, E. | | 登録日 | 2017-10-30 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Integrative Structural Biology Studies of HIV-1 Reverse Transcriptase Binding to a High-Affinity DNA Aptamer
Curr Res Struct Biol, 2020
|
|
6BI4
 
 | | 2.9 Angstrom Resolution Crystal Structure of dTDP-Glucose 4,6-dehydratase (rfbB) from Bacillus anthracis str. Ames in Complex with NAD. | | 分子名称: | NICKEL (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Halavaty, A.S, Kuhn, M, Shuvalova, L, Minasov, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-10-31 | | 公開日 | 2017-11-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Structure of the Bacillus anthracis dTDP-l-rhamnose biosynthetic pathway enzyme: dTDP-alpha-d-glucose 4,6-dehydratase, RfbB.
J.Struct.Biol., 202, 2018
|
|
6EEV
 
 | | Structure of class II HMG-CoA reductase from Delftia acidovorans with mevalonate bound | | 分子名称: | (R)-MEVALONATE, 3-hydroxy-3-methylglutaryl coenzyme A reductase, GLYCEROL, ... | | 著者 | Ragwan, E.R, Arai, E, Kung, Y. | | 登録日 | 2018-08-15 | | 公開日 | 2018-09-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | New Crystallographic Snapshots of Large Domain Movements in Bacterial 3-Hydroxy-3-methylglutaryl Coenzyme A Reductase.
Biochemistry, 57, 2018
|
|
6HAK
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with a double stranded RNA represents the RT transcription initiation complex prior to nucleotide incorporation | | 分子名称: | Gag-Pol polyprotein, MAGNESIUM ION, RNA (5'-R(P*AP*GP*UP*GP*GP*CP*GP*GP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3'), ... | | 著者 | Das, K, Martinez, S.E, Arnold, E. | | 登録日 | 2018-08-07 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.95 Å) | | 主引用文献 | Structure of HIV-1 RT/dsRNA initiation complex prior to nucleotide incorporation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HVM
 
 | |
6HVN
 
 | | CdaA-APO Y187A Mutant | | 分子名称: | CHLORIDE ION, Diadenylate cyclase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | 著者 | Heidemann, J.L, Neumann, P, Ficner, R. | | 登録日 | 2018-10-11 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.234 Å) | | 主引用文献 | Crystal structures of the c-di-AMP-synthesizing enzyme CdaA.
J.Biol.Chem., 294, 2019
|
|
