3EGN
 
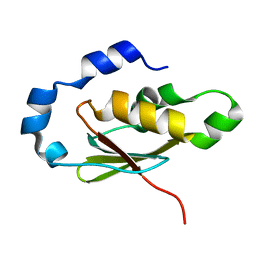 | |
3EX7
 
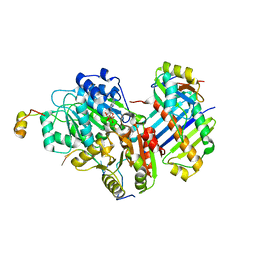 | |
3FEY
 
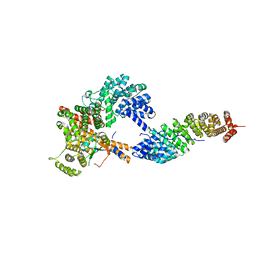 | |
3FEX
 
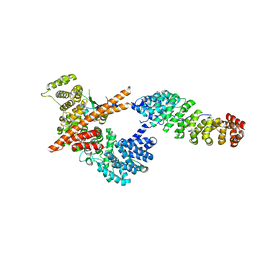 | |
2RQ4
 
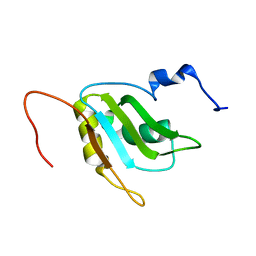 | | Refinement of RNA binding domain 3 in CUG triplet repeat RNA-binding protein 1 | | 分子名称: | CUG-BP- and ETR-3-like factor 1 | | 著者 | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Inoue, M, Terada, T, Kobayashi, N, Shirouzu, M, Kigawa, T, Guntert, P, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-01-19 | | 公開日 | 2009-08-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3
Nucleic Acids Res., 2009
|
|
3G8S
 
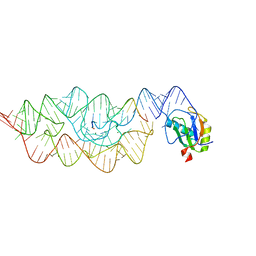 | | Crystal structure of the pre-cleaved Bacillus anthracis glmS ribozyme | | 分子名称: | GLMS RIBOZYME, MAGNESIUM ION, RNA (5'-R(*AP*(A2M)P*GP*CP*GP*CP*CP*AP*GP*AP*AP*CP*U)-3'), ... | | 著者 | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | 登録日 | 2009-02-12 | | 公開日 | 2009-11-03 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
3G8T
 
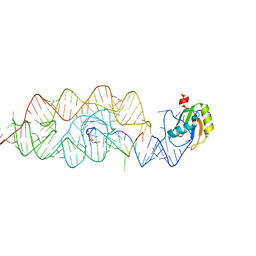 | | Crystal structure of the G33A mutant Bacillus anthracis glmS ribozyme bound to GlcN6P | | 分子名称: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, RNA (5'-R(*AP*(A2M)P*GP*CP*GP*CP*CP*AP*GP*AP*AP*CP*U)-3'), ... | | 著者 | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | 登録日 | 2009-02-12 | | 公開日 | 2009-11-03 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
3G96
 
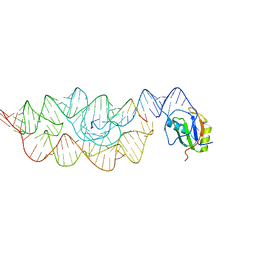 | | Crystal structure of the Bacillus anthracis glmS ribozyme bound to MaN6P | | 分子名称: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-mannopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | 著者 | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | 登録日 | 2009-02-12 | | 公開日 | 2009-11-03 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
3G9C
 
 | | Crystal structure of the product Bacillus anthracis glmS ribozyme | | 分子名称: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | 著者 | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | 登録日 | 2009-02-13 | | 公開日 | 2009-11-03 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
2KG1
 
 | |
2KG0
 
 | |
2KH9
 
 | |
2KHC
 
 | |
2RQC
 
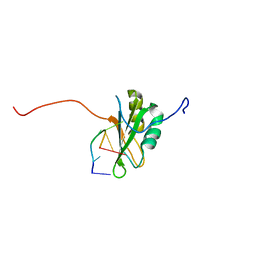 | | Solution Structure of RNA-binding domain 3 of CUGBP1 in complex with RNA (UG)3 | | 分子名称: | 5'-R(*UP*GP*UP*GP*UP*G)-3', CUG-BP- and ETR-3-like factor 1 | | 著者 | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-04-09 | | 公開日 | 2009-08-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3
Nucleic Acids Res., 37, 2009
|
|
3H2V
 
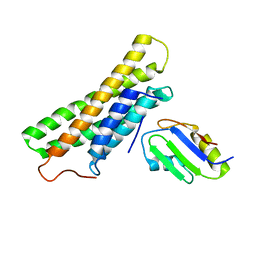 | | Human raver1 RRM1 domain in complex with human vinculin tail domain Vt | | 分子名称: | Raver-1, Vinculin | | 著者 | Lee, J.H, Rangarajan, E.S, Yogesha, S.D, Izard, T. | | 登録日 | 2009-04-14 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Raver1 interactions with vinculin and RNA suggest a feed-forward pathway in directing mRNA to focal adhesions
Structure, 17, 2009
|
|
3H2U
 
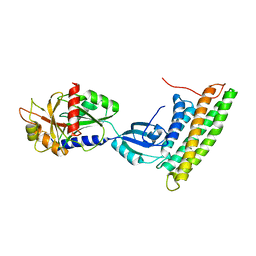 | | Human raver1 RRM1, RRM2, and RRM3 domains in complex with human vinculin tail domain Vt | | 分子名称: | Raver-1, Vinculin | | 著者 | Lee, J.H, Rangarajan, E.S, Yogesha, S.D, Izard, T. | | 登録日 | 2009-04-14 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Raver1 interactions with vinculin and RNA suggest a feed-forward pathway in directing mRNA to focal adhesions
Structure, 17, 2009
|
|
2KI2
 
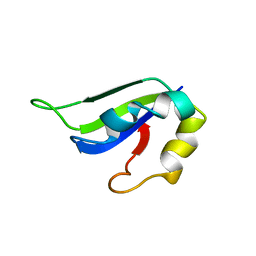 | | Solution Structure of ss-DNA Binding Protein 12RNP2 Precursor, HP0827(O25501_HELPY) form Helicobacter pylori | | 分子名称: | Ss-DNA binding protein 12RNP2 | | 著者 | Ma, C, Lee, J, Kim, J, Park, S, Kwon, A, Lee, B. | | 登録日 | 2009-04-20 | | 公開日 | 2009-10-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of HP0827 (O25501_HELPY) from Helicobacter pylori: model of the possible RNA-binding site
J.Biochem., 146, 2009
|
|
3HHN
 
 | | Crystal structure of class I ligase ribozyme self-ligation product, in complex with U1A RBD | | 分子名称: | Class I ligase ribozyme, self-ligation product, MAGNESIUM ION, ... | | 著者 | Shechner, D.M, Grant, R.A, Bagby, S.C, Bartel, D.P. | | 登録日 | 2009-05-15 | | 公開日 | 2009-11-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.987 Å) | | 主引用文献 | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
3HI9
 
 | |
2KM8
 
 | |
3IIN
 
 | | Plasticity of the kink turn structural motif | | 分子名称: | DNA/RNA (5'-R(*AP*AP*GP*CP*CP*AP*CP*AP*CP*AP*GP*AP*CP*C)-D(P*AP*GP*A)-R(P*CP*GP*GP*CP*C)-3'), DNA/RNA (5'-R(*CP*A)-D(P*T)-3'), Group I intron, ... | | 著者 | Lipchock, S.V, Strobel, S.A, Antonioli, A.H, Cochrane, J.C. | | 登録日 | 2009-08-02 | | 公開日 | 2010-03-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (4.18 Å) | | 主引用文献 | Plasticity of the RNA kink turn structural motif.
Rna, 16, 2010
|
|
2KN4
 
 | |
3IRW
 
 | | Structure of a c-di-GMP riboswitch from V. cholerae | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | 著者 | Smith, K.D. | | 登録日 | 2009-08-24 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis of ligand binding by a c-di-GMP riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3IWN
 
 | | Co-crystal structure of a bacterial c-di-GMP riboswitch | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), C-di-GMP riboswitch, U1 small nuclear ribonucleoprotein A | | 著者 | Kulshina, N, Baird, N.J, Ferre-D'Amare, A.R. | | 登録日 | 2009-09-02 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3K0J
 
 | | Crystal structure of the E. coli ThiM riboswitch in complex with thiamine pyrophosphate and the U1A crystallization module | | 分子名称: | MAGNESIUM ION, RNA (87-MER), THIAMINE DIPHOSPHATE, ... | | 著者 | Kulshina, N, Edwards, T.E, Ferre-D'Amare, A.R. | | 登録日 | 2009-09-24 | | 公開日 | 2009-12-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch.
Rna, 16, 2010
|
|
