8D81
 
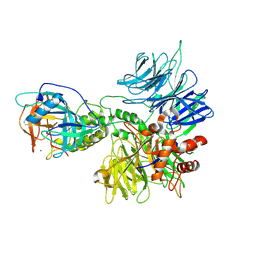 | | Cereblon~DDB1 bound to Pomalidomide | | 分子名称: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | 著者 | Watson, E.R, Lander, G.C. | | 登録日 | 2022-06-07 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
6R8Z
 
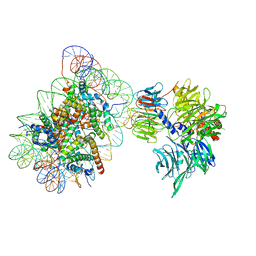 | | Cryo-EM structure of NCP_THF2(-1)-UV-DDB | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6BN8
 
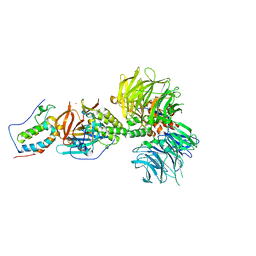 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET55 PROTAC. | | 分子名称: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | 登録日 | 2017-11-16 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.990035 Å) | | 主引用文献 | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6R91
 
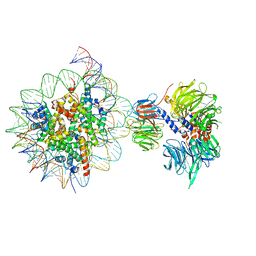 | | Cryo-EM structure of NCP_THF2(-3)-UV-DDB | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
7V7B
 
 | |
6H0G
 
 | | Structure of the DDB1-CRBN-pomalidomide complex bound to ZNF692(ZF4) | | 分子名称: | DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1,DDB1 (DNA damage binding protein 1),DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | 著者 | Bunker, R.D, Petzold, G, Thoma, N.H. | | 登録日 | 2018-07-09 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (4.25 Å) | | 主引用文献 | Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN.
Science, 362, 2018
|
|
6R8Y
 
 | | Cryo-EM structure of NCP-6-4PP(-1)-UV-DDB | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6BN9
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET70 PROTAC | | 分子名称: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | 登録日 | 2017-11-16 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (4.382 Å) | | 主引用文献 | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
8B3G
 
 | | C(N)RL4CSA-UVSSA-E2-ubiquitin complex. | | 分子名称: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | 著者 | Kokic, G, Cramer, P. | | 登録日 | 2022-09-16 | | 公開日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural basis for RNA polymerase II ubiquitylation and inactivation in transcription-coupled repair.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6R90
 
 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class A | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R92
 
 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class B | | 分子名称: | DNA damage-binding protein 1,DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
7ZNN
 
 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and full-length rat STAT2 | | 分子名称: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | 著者 | Lauer, S, Spahn, C.M.T, Schwefel, D. | | 登録日 | 2022-04-21 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
4A0K
 
 | | STRUCTURE OF DDB1-DDB2-CUL4A-RBX1 BOUND TO A 12 BP ABASIC SITE CONTAINING DNA-DUPLEX | | 分子名称: | 12 BP DNA, 12 BP THF CONTAINING DNA, CULLIN-4A, ... | | 著者 | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-09 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (5.93 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
6BNB
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET57 PROTAC | | 分子名称: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Nowak, R.P, DeAngelo, S.L, Buckley, D, Ishoey, M, He, Z, Zhang, T, Bradner, J.E, Fischer, E.S. | | 登録日 | 2017-11-16 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (6.343 Å) | | 主引用文献 | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
4A0L
 
 | | Structure of DDB1-DDB2-CUL4B-RBX1 bound to a 12 bp abasic site containing DNA-duplex | | 分子名称: | 12 BP DNA DUPLEX, 12 BP THF CONTAINING DNA DUPLEX, CULLIN-4B, ... | | 著者 | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-09 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (7.4 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
7OKQ
 
 | | Cryo-EM Structure of the DDB1-DCAF1-CUL4A-RBX1 Complex | | 分子名称: | Cullin-4A, DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, ... | | 著者 | Mohamed, W.I, Schenk, A.D, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2021-05-18 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (8.4 Å) | | 主引用文献 | The CRL4 DCAF1 cullin-RING ubiquitin ligase is activated following a switch in oligomerization state.
Embo J., 40, 2021
|
|
8AJO
 
 | | Negative-stain electron microscopy structure of DDB1-DCAF12-CCT5 | | 分子名称: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, T-complex protein 1 subunit epsilon | | 著者 | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2022-07-28 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (30.6 Å) | | 主引用文献 | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
