2WER
 
 | |
4URM
 
 | | Crystal Structure of Staph GyraseB 24kDa in complex with Kibdelomycin | | 分子名称: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA GYRASE SUBUNIT B | | 著者 | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | 登録日 | 2014-06-30 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
6CJI
 
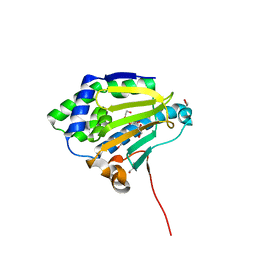 | | Candida albicans Hsp90 nucleotide binding domain | | 分子名称: | 1,2-ETHANEDIOL, Heat shock protein 90 homolog | | 著者 | Hutchinson, A, Loppnau, P, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, STRUCTURAL GENOMICS CONSORTIUM, S.G.C, Pizarro, J.C, Structural Genomics Consortium (SGC) | | 登録日 | 2018-02-26 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus.
Nat Commun, 10, 2019
|
|
6CJR
 
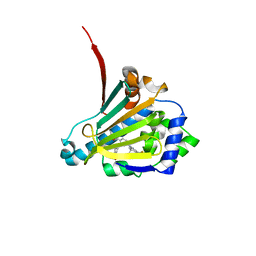 | | Candida albicans Hsp90 nucleotide binding domain in complex with SNX-2112 | | 分子名称: | 1,2-ETHANEDIOL, 4-[6,6-dimethyl-4-oxidanylidene-3-(trifluoromethyl)-5,7-dihydroindazol-1-yl]-2-[(4-oxidanylcyclohexyl)amino]benzamide, Heat shock protein 90 homolog | | 著者 | Kirkpatrick, M.G, Pizarro, J.C. | | 登録日 | 2018-02-26 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus.
Nat Commun, 10, 2019
|
|
2IWS
 
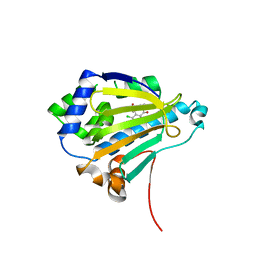 | | Radicicol analogues bound to the ATP site of HSP90 | | 分子名称: | (5Z)-12-CHLORO-13,15-DIHYDROXY-4,7,8,9-TETRAHYDRO-2-BENZOXACYCLOTRIDECINE-1,10(3H,11H)-DIONE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | 著者 | Roe, S.M, Prodromou, C, Pearl, L.H. | | 登録日 | 2006-07-04 | | 公開日 | 2006-11-30 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Inhibition of Hsp90 with Synthetic Macrolactones: Synthesis and Structural and Biological Evaluation of Ring and Conformational Analogs of Radicicol.
Chem.Biol., 13, 2006
|
|
4U7O
 
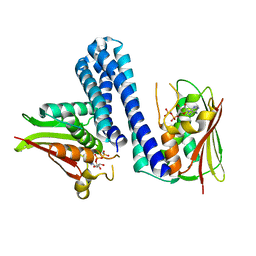 | | Active histidine kinase bound with ATP | | 分子名称: | AMP PHOSPHORAMIDATE, Histidine protein kinase sensor protein | | 著者 | Cai, Y, Hu, X, Sang, J. | | 登録日 | 2014-07-31 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.395 Å) | | 主引用文献 | Conformational dynamics of the essential sensor histidine kinase WalK.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4U7N
 
 | | Inactive structure of histidine kinase | | 分子名称: | Histidine protein kinase sensor protein | | 著者 | Cai, Y, Hu, X, Sang, J. | | 登録日 | 2014-07-31 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Conformational dynamics of the essential sensor histidine kinase WalK.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
2IWU
 
 | | Analogues of radicicol bound to the ATP-binding site of Hsp90 | | 分子名称: | (5E)-12-CHLORO-13,15-DIHYDROXY-4,7,8,9-TETRAHYDRO-2-BENZOXACYCLOTRIDECINE-1,10(3H,11H)-DIONE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | 著者 | Roe, S.M, Prodromou, C, Pearl, L.H. | | 登録日 | 2006-07-04 | | 公開日 | 2006-11-30 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Inhibition of Hsp90 with Synthetic Macrolactones: Synthesis and Structural and Biological Evaluation of Ring and Conformational Analogs of Radicicol.
Chem.Biol., 13, 2006
|
|
2IOR
 
 | | Crystal Structure of the N-terminal Domain of HtpG, the Escherichia coli Hsp90, Bound to ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein htpG, HEXANE-1,6-DIOL, ... | | 著者 | Shiau, A.K, Harris, S.F, Agard, D.A. | | 登録日 | 2006-10-10 | | 公開日 | 2006-11-21 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural Analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements.
Cell(Cambridge,Mass.), 127, 2006
|
|
6Y8O
 
 | | Mycobacterium smegmatis GyrB 22kDa ATPase sub-domain in complex with novobiocin | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, DNA gyrase subunit B, ... | | 著者 | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | 登録日 | 2020-03-05 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
2IWX
 
 | | Analogues of radicicol bound to the ATP-binding site of Hsp90. | | 分子名称: | (5E)-14-CHLORO-15,17-DIHYDROXY-4,7,8,9,10,11-HEXAHYDRO-2-BENZOXACYCLOPENTADECINE-1,12(3H,13H)-DIONE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | 著者 | Roe, S.M, Prodromou, C, Pearl, L.H. | | 登録日 | 2006-07-05 | | 公開日 | 2006-11-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Inhibition of Hsp90 with Synthetic Macrolactones: Synthesis and Structural and Biological Evaluation of Ring and Conformational Analogs of Radicicol.
Chem.Biol., 13, 2006
|
|
6Y8L
 
 | | Mycobacterium thermoresistibile GyrB21 in complex with novobiocin | | 分子名称: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN, ... | | 著者 | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | 登録日 | 2020-03-05 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
6Y8N
 
 | | Mycobacterium thermoresistibile GyrB21 in complex with Redx03863 | | 分子名称: | 1,2-ETHANEDIOL, 4-[(1~{S},5~{R})-6-azanyl-3-azabicyclo[3.1.0]hexan-3-yl]-6-fluoranyl-~{N}-methyl-2-(2-methylpyrimidin-5-yl)oxy-9~{H}-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, ... | | 著者 | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | 登録日 | 2020-03-05 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
6YD9
 
 | | Ecoli GyrB24 with inhibitor 16a | | 分子名称: | 1,2-ETHANEDIOL, DNA gyrase subunit B, N-[6-(3-azanylpropanoylamino)-1,3-benzothiazol-2-yl]-3,4-bis(chloranyl)-5-methyl-1H-pyrrole-2-carboxamide | | 著者 | Barancokova, M, Skok, Z, Benek, O, Cruz, C.D, Tammela, P, Tomasic, T, Zidar, N, Masic, L.P, Zega, A, Stevenson, C.E.M, Mundy, J, Lawson, D.M, Maxwell, A.M, Kikelj, D, Ilas, J. | | 登録日 | 2020-03-20 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Exploring the Chemical Space of Benzothiazole-Based DNA Gyrase B Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
7PTF
 
 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with novobiocin | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA gyrase subunit B, ... | | 著者 | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | 登録日 | 2021-09-27 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PTG
 
 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with EBL2888 | | 分子名称: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B | | 著者 | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | 登録日 | 2021-09-27 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7P2W
 
 | | E.coli GyrB24 with inhibitor LMD92 (EBL2682) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(3-carboxyphenyl)methoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, ... | | 著者 | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | 登録日 | 2021-07-06 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7P2X
 
 | | E.coli GyrB24 with inhibitor KOB20 (EBL2583) | | 分子名称: | (2Z)-2-[[4,5-bis(bromanyl)-1H-pyrrol-2-yl]carbonylimino]-3-(phenylmethyl)-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | 著者 | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Benek, O, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | 登録日 | 2021-07-06 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | E.coli GyrB24 with inhibitor KOB20 (EBL2583)
TO BE PUBLISHED
|
|
7P2M
 
 | | E.coli GyrB24 with inhibitor LMD43 (EBL2560) | | 分子名称: | 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-phenylmethoxy-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | 著者 | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | 登録日 | 2021-07-06 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7P2N
 
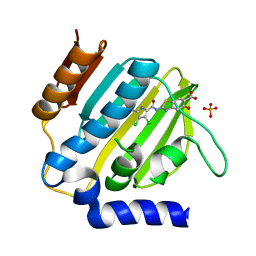 | | E.coli GyrB24 with inhibitor LSJ38 (EBL2684) | | 分子名称: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-5-oxidanyl-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | 著者 | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | 登録日 | 2021-07-06 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Exploring the 5-Substituted 2-Aminobenzothiazole-Based DNA Gyrase B Inhibitors Active against ESKAPE Pathogens.
Acs Omega, 8, 2023
|
|
1KZN
 
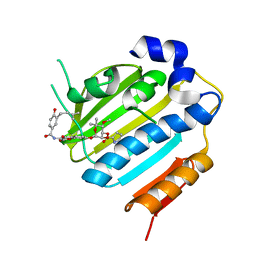 | | Crystal Structure of E. coli 24kDa Domain in Complex with Clorobiocin | | 分子名称: | CLOROBIOCIN, DNA GYRASE SUBUNIT B | | 著者 | Lafitte, D, Lamour, V, Tsvetkov, P.O, Makarov, A.A, Klich, M, Deprez, P, Moras, D, Briand, C, Gilli, R. | | 登録日 | 2002-02-07 | | 公開日 | 2002-06-19 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | DNA gyrase interaction with coumarin-based inhibitors: the role of the hydroxybenzoate isopentenyl moiety and the 5'-methyl group of the noviose.
Biochemistry, 41, 2002
|
|
1S14
 
 | |
1R62
 
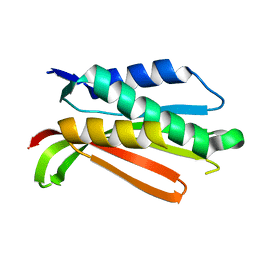 | | Crystal structure of the C-terminal Domain of the Two-Component System Transmitter Protein NRII (NtrB) | | 分子名称: | Nitrogen regulation protein NR(II) | | 著者 | Song, Y, Peisach, D, Pioszak, A.A, Xu, Z, Ninfa, A.J. | | 登録日 | 2003-10-14 | | 公開日 | 2004-06-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of the C-terminal Domain of the Two-Component System Transmitter Protein Nitrogen Regulator II (NRII; NtrB), Regulator of Nitrogen Assimilation in Escherichia coli.
Biochemistry, 43, 2004
|
|
5CTU
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(thiophen-2-yl)thieno[2,3-d]pyrimidin-4(1H)-one, CHLORIDE ION, ... | | 著者 | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | 登録日 | 2015-07-24 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5D6Q
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-{4-[(E)-2-(pyridin-3-yl)ethenyl]-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl}urea, DNA gyrase subunit B, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-12 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
