6GL4
 
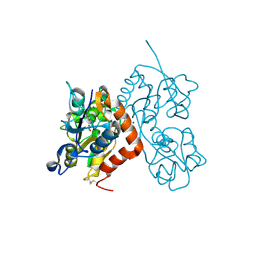 | | Structure of GluA2o ligand-binding domain (S1S2J) in complex with glutamate and sodium bromide at 1.95 A resolution | | 分子名称: | ACETATE ION, BROMIDE ION, GLUTAMIC ACID, ... | | 著者 | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2018-05-22 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.948 Å) | | 主引用文献 | Nanoscale Mobility of the Apo State and TARP Stoichiometry Dictate the Gating Behavior of Alternatively Spliced AMPA Receptors.
Neuron, 102, 2019
|
|
6GIV
 
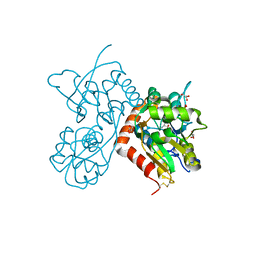 | | Structure of GluA2-N775S ligand-binding domain (S1S2J) in complex with glutamate and Rubidium Bromide at 1.75 A resolution | | 分子名称: | BROMIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | 著者 | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2018-05-15 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Nanoscale Mobility of the Apo State and TARP Stoichiometry Dictate the Gating Behavior of Alternatively Spliced AMPA Receptors.
Neuron, 102, 2019
|
|
7LDD
 
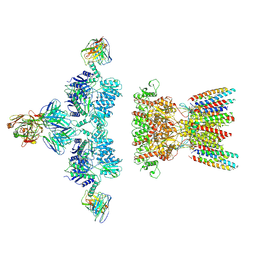 | | native AMPA receptor | | 分子名称: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | 著者 | Yu, J, Rao, P, Gouaux, E. | | 登録日 | 2021-01-13 | | 公開日 | 2021-05-12 | | 最終更新日 | 2021-06-30 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
7LDE
 
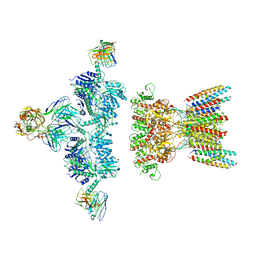 | | native AMPA receptor | | 分子名称: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | 著者 | Yu, J, Rao, P, Gouaux, E. | | 登録日 | 2021-01-13 | | 公開日 | 2021-05-12 | | 最終更新日 | 2021-06-30 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
7LEP
 
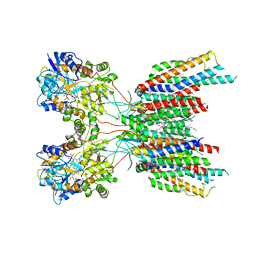 | | The composite LBD-TMD structure combined from all hippocampal AMPAR subtypes at 3.25 Angstrom resolution | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 6-[2-chloro-6-(trifluoromethoxy)phenyl]-1H-benzimidazol-2-ol, DECANE, ... | | 著者 | Yu, J, Rao, P, Gouaux, E. | | 登録日 | 2021-01-14 | | 公開日 | 2021-05-12 | | 最終更新日 | 2021-06-30 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
6FQH
 
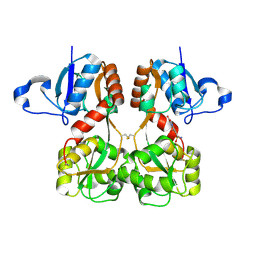 | | GluA2(flop) S729C ligand binding core dimer bound to NBQX at 1.76 Angstrom resolution | | 分子名称: | 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide, Glutamate receptor 2,Glutamate receptor 2 | | 著者 | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | 登録日 | 2018-02-14 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.75940013 Å) | | 主引用文献 | Homomeric GluA2(R) AMPA receptors can conduct when desensitized.
Nat Commun, 10, 2019
|
|
7LZI
 
 | |
7LZ2
 
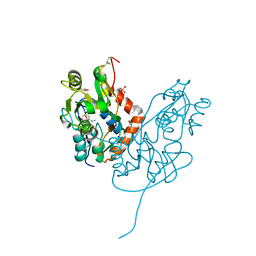 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with methionine | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | 登録日 | 2021-03-08 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
7LZH
 
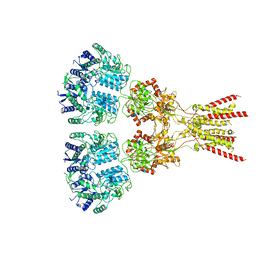 | | Structure of the glutamate receptor-like channel AtGLR3.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | 著者 | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | 登録日 | 2021-03-09 | | 公開日 | 2021-07-28 | | 最終更新日 | 2021-08-18 | | 実験手法 | ELECTRON MICROSCOPY (3.57 Å) | | 主引用文献 | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
7LZ0
 
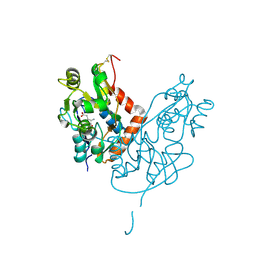 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with glutamate | | 分子名称: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | 著者 | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | 登録日 | 2021-03-08 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
7LZ1
 
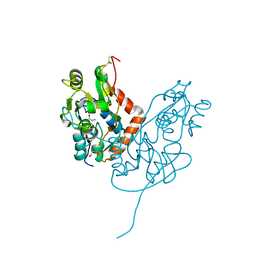 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with serine | | 分子名称: | GLYCEROL, Glutamate receptor 3.4, SERINE, ... | | 著者 | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | 登録日 | 2021-03-08 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
1SYH
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 1.85 A RESOLUTION. | | 分子名称: | (S)-2-AMINO-3-(1,3,5,7-PENTAHYDRO-2,4-DIOXO-CYCLOPENTA[E]PYRIMIDIN-1-YL) PROIONIC ACID, Glutamate receptor 2 | | 著者 | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | 登録日 | 2004-04-01 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
7LVT
 
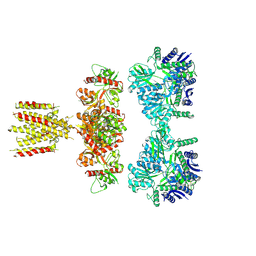 | |
1S9T
 
 | |
1S7Y
 
 | |
1SYI
 
 | | X-RAY STRUCTURE OF THE Y702F MUTANT OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 2.1 A RESOLUTION. | | 分子名称: | (S)-2-AMINO-3-(1,3,5,7-PENTAHYDRO-2,4-DIOXO-CYCLOPENTA[E]PYRIMIDIN-1-YL) PROIONIC ACID, Glutamate receptor 2 | | 著者 | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | 登録日 | 2004-04-01 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
1TXF
 
 | |
1TT1
 
 | |
2QS4
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with LY466195 at 1.58 Angstroms resolution | | 分子名称: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, AMMONIUM ION, GLYCEROL, ... | | 著者 | Alushin, G.M, Jane, D.E, Mayer, M.L. | | 登録日 | 2007-07-30 | | 公開日 | 2008-08-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2QS1
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP315 at 1.80 Angstroms resolution | | 分子名称: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-methyl-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)-4,5-dibromothiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | 著者 | Alushin, G.M, Jane, D.E, Mayer, M.L. | | 登録日 | 2007-07-30 | | 公開日 | 2008-08-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2RC9
 
 | |
2RC8
 
 | |
2V3T
 
 | | Structure of the ligand-binding core of the ionotropic glutamate receptor-like GluRdelta2 in the apo form | | 分子名称: | CALCIUM ION, GLUTAMATE RECEPTOR DELTA-2 SUBUNIT SYNONYM GLURDELTA2, GLUR DELTA-2 | | 著者 | Naur, P, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | 登録日 | 2007-06-22 | | 公開日 | 2007-08-07 | | 最終更新日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Ionotropic Glutamate-Like Receptor {Delta}2 Binds D-Serine and Glycine.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2UXA
 
 | | Crystal structure of the GluR2-flip ligand binding domain, r/g unedited. | | 分子名称: | GLUTAMATE RECEPTOR SUBUNIT GLUR2-FLIP, GLUTAMIC ACID, ZINC ION | | 著者 | Greger, I.H, Akamine, P, Khatri, L, Ziff, E.B. | | 登録日 | 2007-03-27 | | 公開日 | 2007-04-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Developmentally Regulated, Combinatorial RNA Processing Modulates Ampa Receptor Biogenesis.
Neuron, 51, 2006
|
|
2V3U
 
 | | Structure of the ligand-binding core of the ionotropic glutamate receptor-like GluRdelta2 in complex with D-serine | | 分子名称: | CHLORIDE ION, D-SERINE, GLUTAMATE RECEPTOR DELTA-2 SUBUNIT, ... | | 著者 | Naur, P, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | 登録日 | 2007-06-22 | | 公開日 | 2007-08-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Ionotropic Glutamate-Like Receptor {Delta}2 Binds D-Serine and Glycine.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
