7D4F
 
 | | Structure of COVID-19 RNA-dependent RNA polymerase bound to suramin | | 分子名称: | 8-(3-(3-aminobenzamido)-4-methylbenzamido)naphthalene-1,3,5-trisulfonic acid, Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Li, Z, Yin, W, Zhou, Z, Yu, X, Xu, H. | | 登録日 | 2020-09-23 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
9AVQ
 
 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | 著者 | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | 登録日 | 2024-03-04 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
7KOL
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496 inhibitor | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, CHLORIDE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-11-09 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496
to be published
|
|
7CMD
 
 | |
7AQJ
 
 | | Structure of SARS-CoV-2 Main Protease bound to Triglycidyl isocyanurate. | | 分子名称: | 1-[(2~{R})-2-oxidanylpropyl]-3-[[(2~{R})-oxiran-2-yl]methyl]-5-[[(2~{S})-oxiran-2-yl]methyl]-1,3,5-triazinane-2,4,6-trione, 3C-like proteinase, Triglycidyl isocyanurate | | 著者 | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-10-22 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
8DII
 
 | |
7KEH
 
 | | Crystal structure from SARS-CoV-2 NendoU NSP15 | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | 登録日 | 2020-10-10 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7MLF
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) Covalently Bound to Compound C7 | | 分子名称: | 3C-like proteinase, N-(4-tert-butylphenyl)-2-chloro-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]acetamide | | 著者 | Sharon, I, Stille, J, Tjutrins, J, Wang, G, Venegas, F.A, Hennecker, C, Rueda, A.M, Miron, C.E, Pinus, S, Labarre, A, Patrascu, M.B, Vlaho, D, Huot, M, Mittermaier, A.K, Moitessier, N, Schmeing, T.M. | | 登録日 | 2021-04-28 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CL pro covalent inhibitors.
Eur.J.Med.Chem., 229, 2021
|
|
7S70
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI34 | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-4-(butylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Yang, K.S, Sankaran, B, Liu, W.R. | | 登録日 | 2021-09-15 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7KX5
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with noncovalent inhibitor Jun8-76-3A | | 分子名称: | 3C-like proteinase, GLYCEROL, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]furan-2-carboxamide | | 著者 | Sacco, M, Wang, J, Chen, Y. | | 登録日 | 2020-12-03 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Discovery of Di- and Trihaloacetamides as Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity.
J.Am.Chem.Soc., 143, 2021
|
|
6ZMI
 
 | | SARS-CoV-2 Nsp1 bound to the human LYAR-80S ribosome complex | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-02 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
8DK8
 
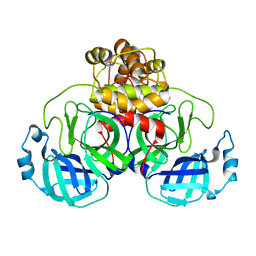 | |
6ZLW
 
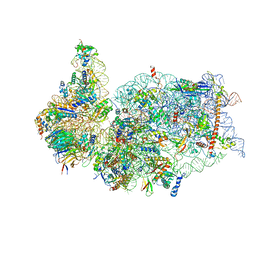 | | SARS-CoV-2 Nsp1 bound to the human 40S ribosomal subunit | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-01 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
7K9P
 
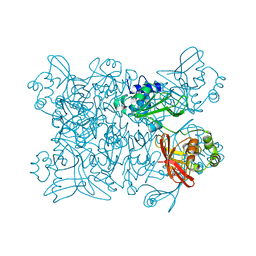 | | Room temperature structure of NSP15 Endoribonuclease from SARS CoV-2 solved using SFX. | | 分子名称: | CITRIC ACID, Uridylate-specific endoribonuclease | | 著者 | Botha, S, Jernigan, R, Chen, J, Coleman, M.A, Frank, M, Grant, T.D, Hansen, D.T, Ketawala, G, Logeswaran, D, Martin-Garcia, J, Nagaratnam, N, Raj, A.L.L.X, Shelby, M, Yang, J.-H, Yung, M.C, Fromme, P. | | 登録日 | 2020-09-29 | | 公開日 | 2020-10-21 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Room-temperature structural studies of SARS-CoV-2 protein NendoU with an X-ray free-electron laser.
Structure, 2022
|
|
8AIV
 
 | | Mpro of SARS COV-2 in complex with the MG-100 inhibitor | | 分子名称: | 3C-like proteinase nsp5, CHLORIDE ION, tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(methylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | 著者 | El Kilani, H, Hilgenfeld, R. | | 登録日 | 2022-07-27 | | 公開日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Main Protease SARS-COV-2 in complex with the inhibitor MG-100
To Be Published
|
|
7DOI
 
 | |
7KF4
 
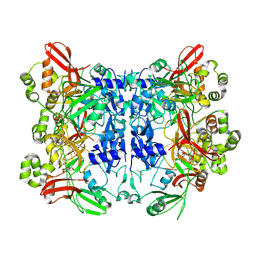 | | Crystal structure from SARS-CoV-2 NendoU NSP15 | | 分子名称: | CITRIC ACID, Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | 登録日 | 2020-10-13 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
9EWM
 
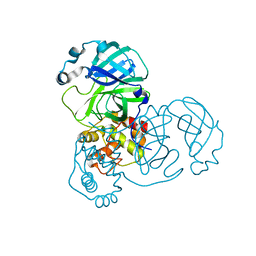 | | Mpro from SARS-CoV-2 with R4Q R298Q double mutations | | 分子名称: | Non-structural protein 11 | | 著者 | Plewka, J, Lis, K, Chykunova, Y, Czarna, A, Kantyka, T, Pyrc, K. | | 登録日 | 2024-04-04 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
8GWM
 
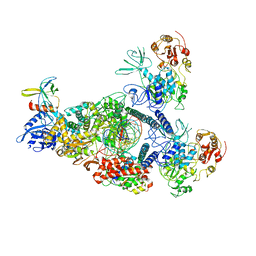 | | SARS-CoV-2 E-RTC bound with MMP-nsp9 and GMPPNP | | 分子名称: | 2'-deoxy-2'-fluoro-2'-methyluridine 5'-(trihydrogen diphosphate), Helicase, Non-structural protein 7, ... | | 著者 | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-17 | | 公開日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
5SMH
 
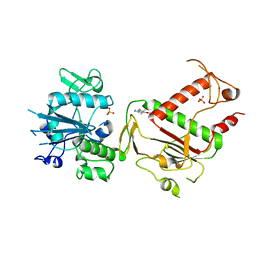 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2856434938 | | 分子名称: | 1-(5-methoxy-1H-indol-3-yl)-N,N-dimethyl-methanamine, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | 著者 | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | 登録日 | 2022-03-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
7CZ4
 
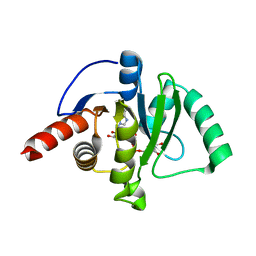 | |
7WZO
 
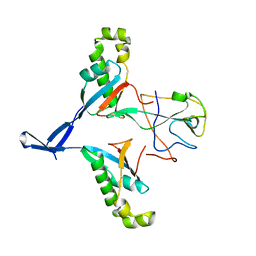 | |
7JIB
 
 | | Room Temperature Crystal Structure of Nsp10/Nsp16 from SARS-CoV-2 with Substrates and Products of 2'-O-methylation of the Cap-1 | | 分子名称: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | 著者 | Wilamowski, M, Minasov, G, Kim, Y, Sherrell, D.A, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-23 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8GWE
 
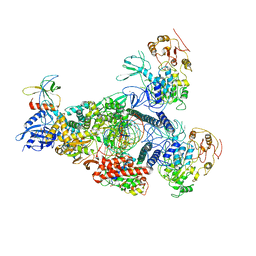 | | SARS-CoV-2 E-RTC complex with RNA-nsp9 and GMPPNP | | 分子名称: | Helicase nsp13, MAGNESIUM ION, Non-structural protein 8, ... | | 著者 | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-16 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8UD3
 
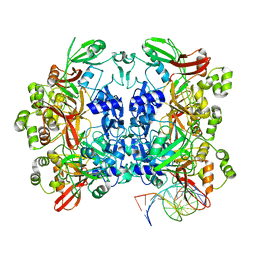 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, consensus form | | 分子名称: | Non-structural protein 15, RNA (35-MER) | | 著者 | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | 登録日 | 2023-09-28 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.67 Å) | | 主引用文献 | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 15, 2024
|
|
