7ZUV
 
 | | Crystal structure of Chlamydomonas reinhardtii chloroplastic sedoheptulose-1,7-bisphosphatase in reducing conditions | | Descriptor: | FBPase domain-containing protein, SULFATE ION | | Authors: | Le Moigne, T, Robert, G.Q, Lemaire, S.D, Henri, J. | | Deposit date: | 2022-05-13 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Characterization of chloroplast ribulose-5-phosphate-3-epimerase from the microalga Chlamydomonas reinhardtii.
Plant Physiol., 194, 2024
|
|
7B2O
 
 | |
6LS5
 
 | | Structure of human liver FBPase complexed with covalent allosteric inhibitor | | Descriptor: | 2-(ethyldisulfanyl)-1,3-benzothiazole, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Yunyuan, H, Rongrong, S, Yixiang, X, Shuaishuai, N, Yanliang, R, Jian, L, Jian, W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Identification of the New Covalent Allosteric Binding Site of Fructose-1,6-bisphosphatase with Disulfiram Derivatives toward Glucose Reduction.
J.Med.Chem., 63, 2020
|
|
7WJV
 
 | | Crystal structure of human liver FBPase complexed with an covalent inhibitor | | Descriptor: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Cao, H, Huang, Y, Ren, Y, Wan, J. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | N -Acylamino Saccharin as an Emerging Cysteine-Directed Covalent Warhead and Its Application in the Identification of Novel FBPase Inhibitors toward Glucose Reduction.
J.Med.Chem., 65, 2022
|
|
4IR8
 
 | | 1.85 Angstrom Crystal Structure of Putative Sedoheptulose-1,7 bisphosphatase from Toxoplasma gondii | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Sedoheptulose-1,7 bisphosphatase, ... | | Authors: | Minasov, G, Ruan, J, Wawrzak, Z, Halavaty, A, Shuvalova, L, Harb, O.S, Ngo, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Crystal Structure of Putative Sedoheptulose-1,7 bisphosphatase from Toxoplasma gondii.
TO BE PUBLISHED
|
|
8XBK
 
 | | Crystal Structure of Human Liver Fructose-1,6-bisphosphatase Complexed with a Covalent Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Fructose-1,6-bisphosphatase 1, MAGNESIUM ION, ... | | Authors: | Cao, H, Zhang, X, Ren, Y, Wan, J. | | Deposit date: | 2023-12-06 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure-Guided Design of Affinity/Covalent-Bond Dual-Driven Inhibitors Targeting the AMP Site of FBPase.
J.Med.Chem., 67, 2024
|
|
7NS5
 
 | | Structure of yeast Fbp1 (Fructose-1,6-bisphosphatase 1) | | Descriptor: | Fructose-1,6-bisphosphatase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Sherpa, D, Chrustowicz, J, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-03-05 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
4MJO
 
 | | Human liver fructose-1,6-bisphosphatase(d-fructose-1,6-bisphosphate, 1-phosphohydrolase) (e.c.3.1.3.11) complexed with the allosteric inhibitor 3 | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-({4-bromo-6-[(methylcarbamoyl)amino]pyridin-2-yl}carbamoyl)-5-(2-methoxyethyl)-4-methylthiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Tetaz, T, Benz, J. | | Deposit date: | 2013-09-04 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of protein-ligand binding constants of a cooperatively regulated tetrameric enzyme using electrospray mass spectrometry.
Acs Chem.Biol., 9, 2014
|
|
3A29
 
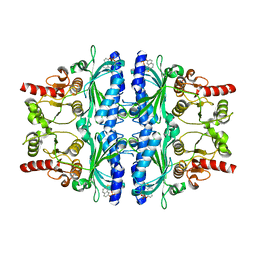 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor | | Descriptor: | 2-amino-4,5-dihydronaphtho[1,2-d][1,3]thiazol-8-yl dihydrogen phosphate, Fructose-1,6-bisphosphatase 1 | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-05-08 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis, SAR, and X-ray structure of tricyclic compounds as potent FBPase inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3FBP
 
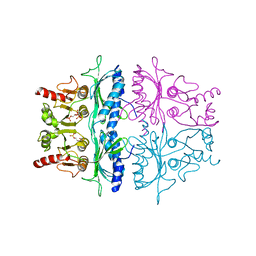 | | STRUCTURE REFINEMENT OF FRUCTOSE-1,6-BISPHOSPHATASE AND ITS FRUCTOSE 2,6-BISPHOSPHATE COMPLEX AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Ke, H, Thorpe, C.M, Seaton, B.A, Marcus, F, Lipscomb, W.N. | | Deposit date: | 1990-06-07 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure refinement of fructose-1,6-bisphosphatase and its fructose 2,6-bisphosphate complex at 2.8 A resolution.
J.Mol.Biol., 212, 1990
|
|
7K74
 
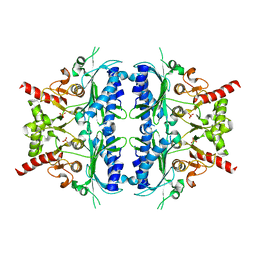 | |
2JJK
 
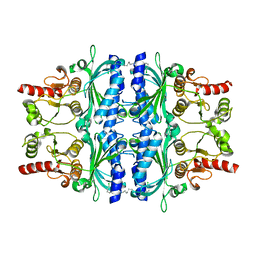 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE -1- PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH A DUAL BINDING AMP SITE INHIBITOR | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N,N'-(heptane-1,7-diyldicarbamoyl)bis(3-chlorobenzenesulfonamide) | | Authors: | Ruf, A, Joseph, C, Benz, J, Fol, B, Tetaz, T, Hebeisen, P. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric Fbpase Inhibitors Gain 10(5) Times in Potency When Simultaneously Binding Two Neighboring AMP Sites.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5ZWK
 
 | | Crystal structure of Human liver fructose-1,6-bisphoaphatase complex with fructose-1,6-bisphophate and AMP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Yunyuan, H, Zeyuan, G, Junjie, Y, Ping, Y, Jian, W. | | Deposit date: | 2018-05-15 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Location of FBPase catalytic metal binding site: a combined experimental and theoretical study
To Be Published
|
|
1BK4
 
 | | CRYSTAL STRUCTURE OF RABBIT LIVER FRUCTOSE-1,6-BISPHOSPHATASE AT 2.3 ANGSTROM RESOLUTION | | Descriptor: | MAGNESIUM ION, PROTEIN (FRUCTOSE-1,6-BISPHOSPHATASE), SULFATE ION | | Authors: | Ghosh, D, Weeks, C.M, Erman, M, Roszak, A.W, Kaiser, R, Jornvall, H. | | Deposit date: | 1998-07-14 | | Release date: | 1998-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of rabbit liver fructose 1,6-bisphosphatase at 2.3 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5IZ1
 
 | | Physcomitrella patens FBPase | | Descriptor: | fructose-1,6-bisphosphatase | | Authors: | Einsle, O, Guetle, D. | | Deposit date: | 2016-03-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6LW2
 
 | | The N-arylsulfonyl-indole-2-carboxamide-based inhibitors against fructose-1,6-bisphosphatase | | Descriptor: | 7-chloranyl-4-[(3-methoxyphenyl)amino]-N-(4-methoxyphenyl)sulfonyl-1-methyl-indole-2-carboxamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2020-02-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of N -Arylsulfonyl-Indole-2-Carboxamide Derivatives as Potent, Selective, and Orally Bioavailable Fructose-1,6-Bisphosphatase Inhibitors-Design, Synthesis, In Vivo Glucose Lowering Effects, and X-ray Crystal Complex Analysis.
J.Med.Chem., 63, 2020
|
|
5K55
 
 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state in complex with fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
5K56
 
 | | Human muscle fructose-1,6-bisphosphatase in active R-state in complex with fructose-1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
3IFC
 
 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in complex with AMP and alpha fructose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase isozyme 2, ... | | Authors: | Kolodziejczyk, R, Zarzycki, M, Jaskolski, M, Dzugaj, A. | | Deposit date: | 2009-07-24 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of E69Q mutant of human muscle fructose-1,6-bisphosphatase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3IFA
 
 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase isozyme 2, GLYCEROL, ... | | Authors: | Kolodziejczyk, R, Zarzycki, M, Jaskolski, M, Dzugaj, A. | | Deposit date: | 2009-07-24 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of E69Q mutant of human muscle fructose-1,6-bisphosphatase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
7WVB
 
 | | Human Fructose-1,6-bisphosphatase 1 mutant R50A in APO R-state | | Descriptor: | Fructose-1,6-bisphosphatase 1 | | Authors: | Chen, Y, Zhang, J, Li, C, Cao, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Human Fructose-1,6-bisphosphatase 1 mutant R50A in APO R-state
To Be Published
|
|
5Q0B
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(4-bromo-3-methyl-1,2-thiazol-5-yl)-3-(3-methylphenyl)sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(4-bromo-3-methyl-1,2-thiazol-5-yl)carbamoyl]-3-methylbenzene-1-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(4-bromo-3-methyl-1,2-thiazol-5-yl)-3-(3-methylphenyl)sulfonylurea
To be published
|
|
3KC0
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor 10b | | Descriptor: | Fructose-1,6-bisphosphatase 1, [(8H-indeno[1,2-d][1,3]thiazol-4-yloxy)methyl]phosphonic acid | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based drug design of tricyclic 8H-indeno[1,2-d][1,3]thiazoles as potent FBPase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1YYZ
 
 | | R-State AMP Complex Reveals Initial Steps of the Quaternary Transition of Fructose-1,6-bisphosphatase | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase, ... | | Authors: | Iancu, C.V, Mukund, S, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | R-state AMP complex reveals initial steps of the quaternary transition of fructose-1,6-bisphosphatase.
J.Biol.Chem., 280, 2005
|
|
3KBZ
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor 6 | | Descriptor: | Fructose-1,6-bisphosphatase 1, {[(2-amino-8H-indeno[1,2-d][1,3]thiazol-4-yl)oxy]methyl}phosphonic acid | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based drug design of tricyclic 8H-indeno[1,2-d][1,3]thiazoles as potent FBPase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
