7LRQ
 
 | | Crystal structure of human SFPQ/NONO heterodimer, conserved DBHS region | | Descriptor: | CHLORIDE ION, Non-POU domain-containing octamer-binding protein, Splicing factor, ... | | Authors: | Marshall, A.C, Bond, C.S, Mohnen, I. | | Deposit date: | 2021-02-17 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Paraspeckle subnuclear bodies depend on dynamic heterodimerisation of DBHS RNA-binding proteins via their structured domains.
J.Biol.Chem., 298, 2022
|
|
8I1I
 
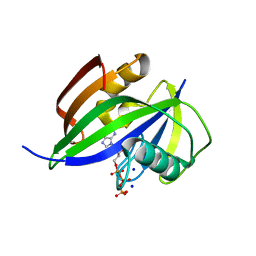 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP at pH 7.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1E
 
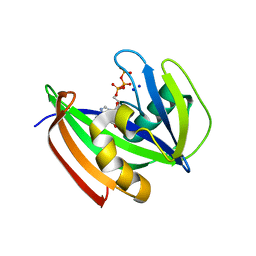 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 8.0 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I18
 
 | |
8I1H
 
 | |
8I1J
 
 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP at pH 9.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1G
 
 | |
8I8T
 
 | |
8I1F
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 8.6 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I19
 
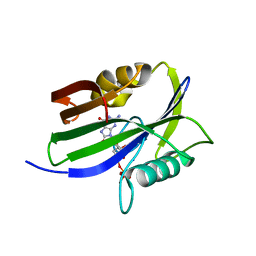 | |
8I8S
 
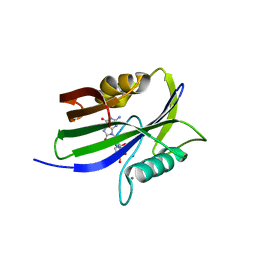 | |
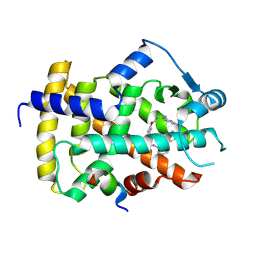
9CWN
 
 | |
8IGL
 
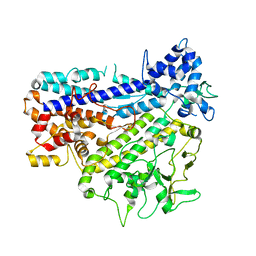 | |
6HCK
 
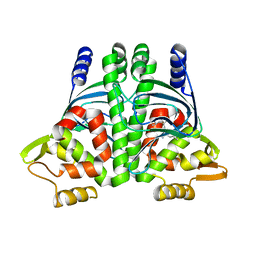 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with dipeptide Leu-Leu | | Descriptor: | LEUCINE, Listeriolysin regulatory protein, SODIUM ION | | Authors: | Grundstrom, C, Oelker, M, Krypotou, E, Scortti, M, Luisi, B.F, Vazquez-Boland, J, Sauer-Eriksson, A.E. | | Deposit date: | 2018-08-15 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Control of Bacterial Virulence through the Peptide Signature of the Habitat.
Cell Rep, 26, 2019
|
|
6HRX
 
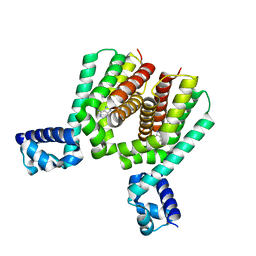 | | EthR2 in complex with compound 2 (BDM72201) | | Descriptor: | 8-propan-2-ylsulfanyl-7~{H}-purin-6-amine, Probable transcriptional regulatory protein | | Authors: | Wintjens, R, Wohlkonig, A, Tanina, A. | | Deposit date: | 2018-09-28 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A fragment-based approach towards the discovery of N-substituted tropinones as inhibitors of Mycobacterium tuberculosis transcriptional regulator EthR2.
Eur J Med Chem, 167, 2019
|
|
9GBK
 
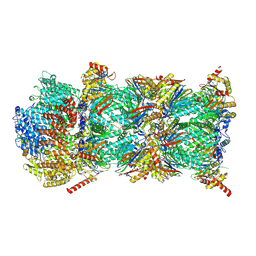 | | Blm10-20S proteasome complex from pre1-1 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome activator BLM10, Proteasome subunit alpha type-1, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-07-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis.
Life Sci Alliance, 7, 2024
|
|
9ETK
 
 | |
4XC5
 
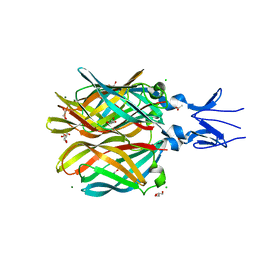 | | CRYSTAL STRUCTURE OF THE T1L REOVIRUS ATTACHMENT PROTEIN SIGMA1 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Reiss, K, Stehle, T. | | Deposit date: | 2014-12-17 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Serotype 1 Reovirus Attachment Protein sigma 1 in Complex with Junctional Adhesion Molecule A Reveals a Conserved Serotype-Independent Binding Epitope.
J.Virol., 89, 2015
|
|
6HS0
 
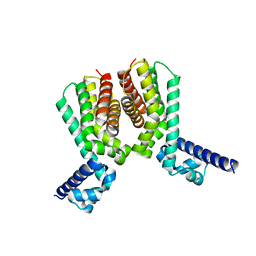 | | EthR2 in complex with compound 5 (BDM71847) | | Descriptor: | 1-[(3-chlorophenyl)methyl]piperazine, Probable transcriptional regulatory protein | | Authors: | Wintjens, R, Wohlkonig, A, Tanina, A. | | Deposit date: | 2018-09-28 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A fragment-based approach towards the discovery of N-substituted tropinones as inhibitors of Mycobacterium tuberculosis transcriptional regulator EthR2.
Eur J Med Chem, 167, 2019
|
|
2ZK1
 
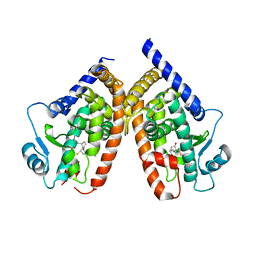 | | Human peroxisome proliferator-activated receptor gamma ligand binding domain complexed with 15-deoxy-delta12,14-prostaglandin J2 | | Descriptor: | (5E,14E)-11-oxoprosta-5,9,12,14-tetraen-1-oic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Waku, T, Shiraki, T, Oyama, T, Fujimoto, Y, Morikawa, K. | | Deposit date: | 2008-03-12 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural insight into PPARgamma activation through covalent modification with endogenous fatty acids
J.Mol.Biol., 385, 2009
|
|
2ZMI
 
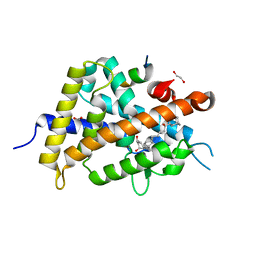 | | Crystal Structure of Rat Vitamin D Receptor Bound to Adamantyl Vitamin D Analogs: Structural Basis for Vitamin D Receptor Antagonism and/or Partial Agonism | | Descriptor: | (1R,3R,7E,17beta)-17-{(1S,2E,5R)-5-hydroxy-1-methyl-5-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]pent-2-en-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Nakabayashi, M, Yamada, S, Tanaka, T, Igarashi, M, Yoshimoto, N, Ikura, T, Ito, N, Makishima, M, Tokiwa, H, DeLuca, H.F, Shimizu, M. | | Deposit date: | 2008-04-19 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of rat vitamin d receptor bound to adamantyl vitamin d analogs: structural basis for vitamin d receptor antagonism and partial agonism
J.Med.Chem., 51, 2008
|
|
2ZK4
 
 | | Human peroxisome proliferator-activated receptor gamma ligand binding domain complexed with 15-oxo-eicosatetraenoic acid | | Descriptor: | (5E,8E,11Z,13E)-15-oxoicosa-5,8,11,13-tetraenoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Waku, T, Shiraki, T, Oyama, T, Fujimoto, Y, Morikawa, K. | | Deposit date: | 2008-03-12 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural insight into PPARgamma activation through covalent modification with endogenous fatty acids
J.Mol.Biol., 385, 2009
|
|
2ZK6
 
 | | Human peroxisome proliferator-activated receptor gamma ligand binding domain complexed with C8-BODIPY | | Descriptor: | Peroxisome proliferator-activated receptor gamma, difluoro(5-{2-[(5-octyl-1H-pyrrol-2-yl-kappaN)methylidene]-2H-pyrrol-5-yl-kappaN}pentanoato)boron | | Authors: | Waku, T, Shiraki, T, Oyama, T, Fujimoto, Y, Morikawa, K. | | Deposit date: | 2008-03-12 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The nuclear receptor PPARgamma individually responds to serotonin- and fatty acid-metabolites
Embo J., 29, 2010
|
|
9D8A
 
 | |
2ZMJ
 
 | | Crystal Structure of Rat Vitamin D Receptor Bound to Adamantyl Vitamin D Analogs: Structural Basis for Vitamin D Receptor Antagonism and/or Partial Agonism | | Descriptor: | (1R,3R,7E,17beta)-17-{(1S,2E,5R)-5-hydroxy-1-methyl-6-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]hex-2-en-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Yamada, S, Tanaka, T, Igarashi, M, Yoshimoto, N, Ikura, T, Ito, N, Makishima, M, Tokiwa, H, DeLuca, H.F, Shimizu, M. | | Deposit date: | 2008-04-19 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of rat vitamin d receptor bound to adamantyl vitamin d analogs: structural basis for vitamin d receptor antagonism and partial agonism
J.Med.Chem., 51, 2008
|
|
