5WWS
 
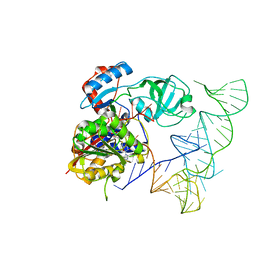 | | Crystal structure of human NSun6/tRNA/SAM | | Descriptor: | Putative methyltransferase NSUN6, S-ADENOSYLMETHIONINE, tRNA | | Authors: | Liu, R.J, Long, T, Wang, E.D. | | Deposit date: | 2017-01-04 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.247 Å) | | Cite: | Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6
Nucleic Acids Res., 45, 2017
|
|
1K74
 
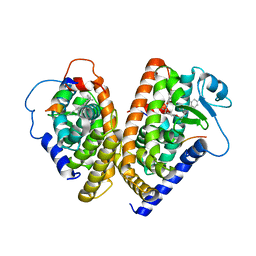 | | The 2.3 Angstrom resolution crystal structure of the heterodimer of the human PPARgamma and RXRalpha ligand binding domains respectively bound with GW409544 and 9-cis retinoic acid and co-activator peptides. | | Descriptor: | (9cis)-retinoic acid, 2-(1-METHYL-3-OXO-3-PHENYL-PROPYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator activated receptor gamma, ... | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Moore, L.B, Collins, J.L, Oplinger, J.A, Kliewer, S.A, Gampe Jr, R.T, McKee, D.D, Moore, J.T, Willson, T.M. | | Deposit date: | 2001-10-18 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7LOE
 
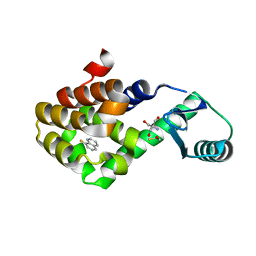 | | T4 lysozyme mutant L99A in complex with 1-fluoranylnaphthalene | | Descriptor: | 1-fluoranylnaphthalene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1APA
 
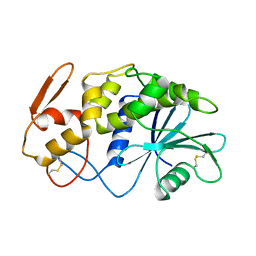 | | X-RAY STRUCTURE OF A POKEWEED ANTIVIRAL PROTEIN, CODED BY A NEW GENOMIC CLONE, AT 0.23 NM RESOLUTION. A MODEL STRUCTURE PROVIDES A SUITABLE ELECTROSTATIC FIELD FOR SUBSTRATE BINDING. | | Descriptor: | POKEWEED ANTIVIRAL PROTEIN | | Authors: | Ago, H, Kataoka, J, Tsuge, H, Habuka, N, Inagaki, E, Noma, M, Miyano, M. | | Deposit date: | 1993-09-21 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of a pokeweed antiviral protein, coded by a new genomic clone, at 0.23 nm resolution. A model structure provides a suitable electrostatic field for substrate binding.
Eur.J.Biochem., 225, 1994
|
|
1TRA
 
 | | RESTRAINED REFINEMENT OF THE MONOCLINIC FORM OF YEAST PHENYLALANINE TRANSFER RNA. TEMPERATURE FACTORS AND DYNAMICS, COORDINATED WATERS, AND BASE-PAIR PROPELLER TWIST ANGLES | | Descriptor: | MAGNESIUM ION, TRNAPHE | | Authors: | Westhof, E, Sundaralingam, M. | | Deposit date: | 1986-05-16 | | Release date: | 1986-07-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Restrained refinement of the monoclinic form of yeast phenylalanine transfer RNA. Temperature factors and dynamics, coordinated waters, and base-pair propeller twist angles.
Biochemistry, 25, 1986
|
|
7LOA
 
 | | T4 lysozyme mutant L99A in complex with 3-fluoroiodobenzene | | Descriptor: | 1-fluoranyl-3-iodanyl-benzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-02-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1OKO
 
 | | Crystal structure of Pseudomonas Aeruginosa Lectin 1 complexed with galactose at 1.6 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, PA-I GALACTOPHILIC LECTIN, ... | | Authors: | Cioci, G, Mitchell, E, Gautier, C, Wimmerova, M, Perez, S, Gilboa-Garber, N, Imberty, A. | | Deposit date: | 2003-07-28 | | Release date: | 2003-12-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of Calcium and Galactose Recognition by the Lectin Pa-Il of Pseudomonas Aeruginosa
FEBS Lett., 555, 2003
|
|
7LOF
 
 | | T4 lysozyme mutant L99A in complex with 2-butylthiophene | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-butylthiophene, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1KAF
 
 | | DNA Binding Domain Of The Phage T4 Transcription Factor MotA (AA105-211) | | Descriptor: | Transcription regulatory protein MOTA | | Authors: | Li, N, Sickmier, E.A, Zhang, R, Joachimiak, A, White, S.W. | | Deposit date: | 2001-11-01 | | Release date: | 2001-11-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The MotA transcription factor from bacteriophage T4 contains a novel DNA-binding domain: the 'double wing' motif.
Mol.Microbiol., 43, 2002
|
|
7WLS
 
 | | Crystal structure of the multidrug efflux transporter BpeB from Burkholderia pseudomallei | | Descriptor: | Efflux pump membrane transporter, TETRAETHYLENE GLYCOL, UNDECYL-MALTOSIDE | | Authors: | Kato, T, Hung, L.-W, Yamashita, E, Okada, U, Terwilliger, T.C, Murakami, S. | | Deposit date: | 2022-01-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structures of multidrug efflux transporters from Burkholderia pseudomallei suggest details of transport mechanism.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1BYY
 
 | | SODIUM CHANNEL IIA INACTIVATION GATE | | Descriptor: | PROTEIN (SODIUM CHANNEL ALPHA-SUBUNIT) | | Authors: | Rohl, C.A, Boeckman, F.A, Baker, C, Scheuer, T, Catterall, W.A, Klevit, R.E. | | Deposit date: | 1998-10-21 | | Release date: | 1999-10-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the sodium channel inactivation gate.
Biochemistry, 38, 1999
|
|
7LOD
 
 | | T4 lysozyme mutant L99A in complex with 1-fluoranyl-4-iodanyl-benzene | | Descriptor: | 1-fluoranyl-4-iodanyl-benzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1BOQ
 
 | | PRO REGION C-TERMINUS: PROTEASE ACTIVE SITE INTERACTIONS ARE CRITICAL IN CATALYZING THE FOLDING OF ALPHA-LYTIC PROTEASE | | Descriptor: | PROTEIN (ALPHA-LYTIC PROTEASE), SULFATE ION | | Authors: | Peters, R.J, Shiau, A.K, Sohl, J.L, Anderson, D.E, Tang, G, Silen, J.L, Agard, D.A. | | Deposit date: | 1998-08-05 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pro region C-terminus:protease active site interactions are critical in catalyzing the folding of alpha-lytic protease.
Biochemistry, 37, 1998
|
|
4LMQ
 
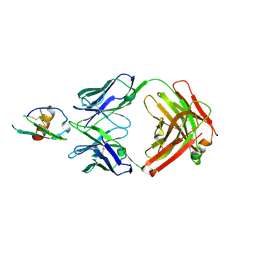 | | Development and Preclinical Characterization of a Humanized Antibody Targeting CXCL12 | | Descriptor: | Stromal cell-derived factor 1, hu30D8 Fab heavy chain, hu30D8 Fab light chain | | Authors: | Zhong, Z, Wang, J, Li, B, Xiang, H, Ultsch, M, Coons, M, Wong, T, Chiang, N.Y, Clark, S, Clark, R, Quintana, L, Gribling, P, Suto, E, Barck, K, Corpuz, R, Yao, J, Takkar, R, Lee, W.P, Damico-Beyer, L.A, Carano, R.D, Adams, C, Kelley, R.F, Wang, W, Ferrara, N. | | Deposit date: | 2013-07-10 | | Release date: | 2013-08-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | Development and Preclinical Characterization of a Humanized Antibody Targeting CXCL12.
Clin.Cancer Res., 19, 2013
|
|
1Z9O
 
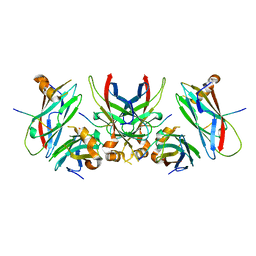 | | 1.9 Angstrom Crystal Structure of the Rat VAP-A MSP Homology Domain in Complex with the Rat ORP1 FFAT Motif | | Descriptor: | Oxysterol binding protein, Vesicle-associated membrane protein-associated protein A | | Authors: | Kaiser, S.E, Brickner, J.H, Reilein, A.R, Fenn, T.D, Walter, P, Brunger, A.T. | | Deposit date: | 2005-04-03 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of FFAT motif-mediated ER targeting
Structure, 13, 2005
|
|
1BNX
 
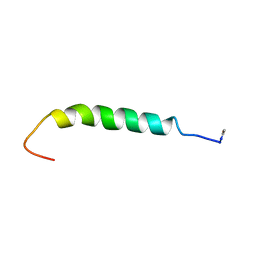 | | STRUCTURAL STUDIES ON THE EFFECTS OF THE DELETION IN THE RED CELL ANION EXCHANGER (BAND3, AE1) ASSOCIATED WITH SOUTH EAST ASIAN OVALOCYTOSIS. | | Descriptor: | PROTEIN (BAND 3) | | Authors: | Chambers, E.J, Bloomberg, G.B, Ring, S.M, Tanner, M.J.A. | | Deposit date: | 1998-07-30 | | Release date: | 1998-08-05 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Studies on the structure of a transmembrane region and a cytoplasmic loop of the human red cell anion exchanger (band 3, AE1).
Biochem.Soc.Trans., 26, 1998
|
|
2I3F
 
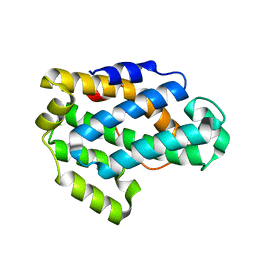 | | Crystal Structure of a Glycolipid transfer-like protein from Galdieria sulphuraria | | Descriptor: | glycolipid transfer-like protein | | Authors: | McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-18 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal Structure of a Glycolipid transfer-like protein from Galdieria sulphuraria
To be Published
|
|
5O2X
 
 | | Extended catalytic domain of H. jecorina LPMO9A a.k.a EG4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Glycoside hydrolase family 61, ... | | Authors: | Hansson, H, Karkehabadi, S, Mikkelsen, N.E, Sandgren, M, Kelemen, B, Kaper, T. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | High-resolution structure of a lytic polysaccharide monooxygenase from Hypocrea jecorina reveals a predicted linker as an integral part of the catalytic domain.
J. Biol. Chem., 292, 2017
|
|
5FRN
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ419 (compound 4c) | | Descriptor: | 4-azanyl-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-6-(5-oxidanylpentyl)-1,8-naphthyridine-3-carboxamide, 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP *TP*CP*GP*CP*A)-3', 5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP *AP*CP*A)-3', ... | | Authors: | Maskell, D.P, Pye, V.E, Cherepanov, P. | | Deposit date: | 2015-12-18 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | HIV-1 Integrase Strand Transfer Inhibitors with Reduced Susceptibility to Drug Resistant Mutant Integrases.
Acs Chem.Biol., 11, 2016
|
|
4P7S
 
 | |
4PIW
 
 | | Crystal structure of sugar aminotransferase WecE from Escherichia coli K-12 | | Descriptor: | TDP-4-keto-6-deoxy-D-glucose transaminase family protein | | Authors: | Wang, F, Xu, W, Helmich, K.E, Singh, S, Yennamalli, R.M, Miller, M.D, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-09 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of sugar aminotransferase WecE from Escherichia coli K-12
To Be Published
|
|
6B5I
 
 | |
1R5T
 
 | | The Crystal Structure of Cytidine Deaminase CDD1, an Orphan C to U editase from Yeast | | Descriptor: | Cytidine deaminase, ZINC ION | | Authors: | Xie, K, Sowden, M.P, Dance, G.S.C, Torelli, A.T, Smith, H.C, Wedekind, J.E. | | Deposit date: | 2003-10-13 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of a yeast RNA-editing deaminase provides insight into the fold and function of activation-induced deaminase and APOBEC-1.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
8DOA
 
 | |
1BW7
 
 | |
