6YVK
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 0.71 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6QJK
 
 | |
6YVL
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 1.42 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YVM
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 2.13 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YVN
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 2.84 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
1BPD
 
 | | CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM | | Descriptor: | DNA POLYMERASE BETA, PHOSPHATE ION | | Authors: | Sawaya, M.R, Pelletier, H, Kumar, A, Wilson, S.H, Kraut, J. | | Deposit date: | 1994-04-12 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism.
Science, 264, 1994
|
|
6YWU
 
 | | Human OMPD-domain of UMPS (K314AcK) in complex with UMP at 1.1 Angstroms resolution | | Descriptor: | GLYCEROL, SULFATE ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-30 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YWT
 
 | |
6YVO
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 3.55 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
1BDV
 
 | | ARC FV10 COCRYSTAL | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*AP*TP*AP*GP*AP*AP*GP*CP*AP*CP*TP*CP*TP*AP*CP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*GP*TP*AP*GP*AP*GP*TP*GP*CP*TP*TP*CP*TP*AP*TP*CP*AP*T)-3'), PROTEIN (ARC FV10 REPRESSOR) | | Authors: | Schildbach, J.F, Karzai, A.W, Raumann, B.E, Sauer, R.T. | | Deposit date: | 1998-05-11 | | Release date: | 1999-01-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Origins of DNA-binding specificity: role of protein contacts with the DNA backbone.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1APN
 
 | |
1B23
 
 | | E. coli cysteinyl-tRNA and T. aquaticus elongation factor EF-TU:GTP ternary complex | | Descriptor: | CYSTEINE, CYSTEINYL TRNA, ELONGATION FACTOR TU, ... | | Authors: | Nissen, P, Kjeldgaard, M, Thirup, S, Nyborg, J. | | Deposit date: | 1998-12-04 | | Release date: | 1998-12-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA.
Structure Fold.Des., 7, 1999
|
|
1B56
 
 | | HUMAN RECOMBINANT EPIDERMAL FATTY ACID BINDING PROTEIN | | Descriptor: | FATTY ACID BINDING PROTEIN, PALMITIC ACID | | Authors: | Van Tilbeurgh, H, Hohoff, C, Borchers, T, Spener, F. | | Deposit date: | 1999-01-12 | | Release date: | 1999-10-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Expression, purification, and crystal structure determination of recombinant human epidermal-type fatty acid binding protein.
Biochemistry, 38, 1999
|
|
6ZS5
 
 | | 3.5 A cryo-EM structure of human uromodulin filament core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
1ARO
 
 | | T7 RNA POLYMERASE COMPLEXED WITH T7 LYSOZYME | | Descriptor: | MERCURY (II) ION, T7 LYSOZYME, T7 RNA POLYMERASE | | Authors: | Steitz, T, Jeruzalmi, D. | | Deposit date: | 1997-08-08 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of T7 RNA polymerase complexed to the transcriptional inhibitor T7 lysozyme.
EMBO J., 17, 1998
|
|
1AR6
 
 | | P1/MAHONEY POLIOVIRUS, DOUBLE MUTANT V1160I +P1095S | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
5K6B
 
 | | Crystal structure of prefusion-stabilized RSV F single-chain 9 DS-Cav1 variant. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, SULFATE ION | | Authors: | Joyce, M.G, Zhang, B, Rundlet, E.J, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
1AVM
 
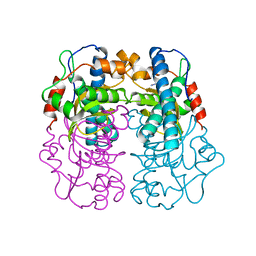 | |
1AXK
 
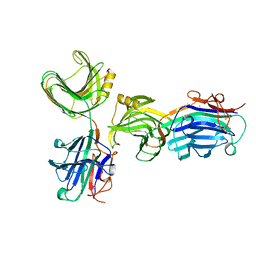 | | ENGINEERED BACILLUS BIFUNCTIONAL ENZYME GLUXYN-1 | | Descriptor: | CALCIUM ION, GLUXYN-1 | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-10-16 | | Release date: | 1999-05-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of the Bacillus hybrid enzyme GluXyn-1: native-like jellyroll fold preserved after insertion of autonomous globular domain.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1AV6
 
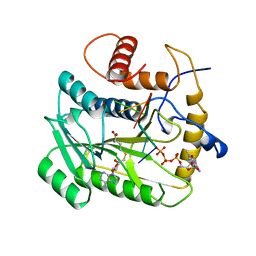 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M7G CAPPED RNA HEXAMER AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, RNA (5'-R(*GP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1997-09-26 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for sequence-nonspecific recognition of 5'-capped mRNA by a cap-modifying enzyme.
Mol.Cell, 1, 1998
|
|
7FQI
 
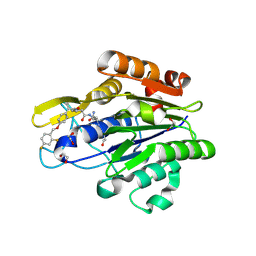 | | Crystal Structure of human Legumain in complex with (2S)-N-[(1S)-3-amino-1-cyano-3-oxopropyl]-1-[1-[4-[(2,4-difluorophenyl)methoxy]phenyl]cyclopropanecarbonyl]pyrrolidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Legumain, ... | | Authors: | Ehler, A, Benz, J, Bartels, B, Rudolph, M.G. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of a human Legumain complex
To be published
|
|
7FQJ
 
 | | Crystal Structure of human Legumain in complex with (2S)-N-[(1S)-3-amino-1-cyano-3-oxopropyl]-1-[1-[4-[(2,4-difluorophenyl)methoxy]phenyl]cyclopropanecarbonyl]pyrrolidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ehler, A, Benz, J, Bartels, B, Rudolph, M.G. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a human Legumain complex
To be published
|
|
7FQK
 
 | | Crystal Structure of human Legumain in complex with (2S)-N-[(3S)-5-amino-1-(1,3-oxazol-2-yl)-5-oxopent-1-yn-3-yl]-1-[1-[4-(trifluoromethoxy)phenyl]cyclopropanecarbonyl]pyrrolidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ehler, A, Benz, J, Bartels, B, Rudolph, M.G. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of a human Legumain complex
To be published
|
|
7FQL
 
 | | Crystal Structure of human Legumain in complex with (2S)-N-[(1S)-3-amino-1-cyano-3-oxopropyl]-1-[1-[4-[(2,4-difluorophenyl)methoxy]phenyl]cyclopropanecarbonyl]pyrrolidine-2-carboxamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Legumain, ... | | Authors: | Ehler, A, Benz, J, Bartels, B, Rudolph, M.G. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal Structure of a human Legumain complex
To be published
|
|
1B05
 
 | |
