2IL4
 
 | | Crystal structure of At1g77540-Coenzyme A Complex | | Descriptor: | COENZYME A, Protein At1g77540 | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
3BFF
 
 | | class A beta-lactamase SED-G238C complexed with faropenem | | Descriptor: | (2R)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-5-[(2R)-oxolan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylic acid, (5R,6S)-6-(1-hydroxyethyl)-7-oxo-3-[(2R)-oxolan-2-yl]-4-thia-1-azabicyclo[3.2.0]hept-2-ene-2-carboxylic acid, Class A beta-lactamase Sed1, ... | | Authors: | Pernot, L, Petrella, S, Sougakoff, W. | | Deposit date: | 2007-11-21 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | acyl-intermediate structures of the class A beta-lactamase SED-G238C
To be Published
|
|
1L3A
 
 | |
3GDJ
 
 | |
3B1D
 
 | | Crystal structure of betaC-S lyase from Streptococcus anginosus in complex with L-serine: External aldimine form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BetaC-S lyase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Kezuka, Y, Yoshida, Y, Nonaka, T. | | Deposit date: | 2011-06-29 | | Release date: | 2012-06-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural insights into catalysis by beta C-S lyase from Streptococcus anginosus
Proteins, 80, 2012
|
|
1RB7
 
 | |
1LQM
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Varshney, U, Vijayan, M. | | Deposit date: | 2002-05-10 | | Release date: | 2002-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1R1Q
 
 | |
1RF9
 
 | | Crystal structure of cytochrome P450-cam with a fluorescent probe D-4-AD (Adamantane-1-carboxylic acid-5-dimethylamino-naphthalene-1-sulfonylamino-butyl-amide) | | Descriptor: | 1,2-ETHANEDIOL, ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-BUTYL-AMIDE, Cytochrome P450-cam, ... | | Authors: | Hays, A.-M.A, Dunn, A.R, Gray, H.B, Stout, C.D, Goodin, D.B. | | Deposit date: | 2003-11-07 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational States of Cytochrome P450cam Revealed by Trapping of synthetic Molecular Wires
J.Mol.Biol., 344, 2004
|
|
1LHZ
 
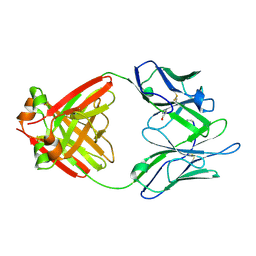 | | Structure of a Human Bence-Jones Dimer Crystallized in U.S. Space Shuttle Mission STS-95: 293K | | Descriptor: | IMMUNOGLOBULIN LAMBDA LIGHT CHAIN | | Authors: | Terzyan, S.S, DeWitt, C.R, Ramsland, P.A, Bourne, P.C, Edmundson, A.B. | | Deposit date: | 2002-04-17 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparison of the three-dimensional structures of a human Bence-Jones dimer
crystallized on Earth and aboard US Space Shuttle Mission STS-95
J.MOL.RECOG., 16, 2003
|
|
2XR9
 
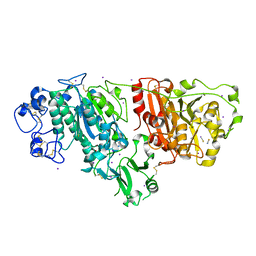 | | Crystal structure of Autotaxin (ENPP2) | | Descriptor: | CALCIUM ION, ECTONUCLEOTIDE PYROPHOSPHATASE/PHOSPHODIESTERASE FAMILY MEMBER 2, IODIDE ION, ... | | Authors: | Kamtekar, S, Hausmann, J, Day, J.E, Christodoulou, E, Perrakis, A. | | Deposit date: | 2010-09-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of substrate discrimination and integrin binding by autotaxin.
Nat. Struct. Mol. Biol., 18, 2011
|
|
2XMH
 
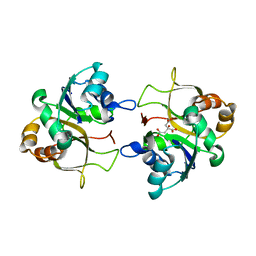 | | The X-ray structure of CTP:inositol-1-phosphate cytidylyltransferase from Archaeoglobus fulgidus | | Descriptor: | CITRATE ANION, CTP-INOSITOL-1-PHOSPHATE CYTIDYLYLTRANSFERASE | | Authors: | Brito, J.A, Borges, N, Vonrhein, C, Santos, H, Archer, M. | | Deposit date: | 2010-07-27 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Archaeoglobus fulgidus CTP:inositol-1-phosphate cytidylyltransferase, a key enzyme for di-myo-inositol-phosphate synthesis in (hyper)thermophiles.
J. Bacteriol., 193, 2011
|
|
3BBW
 
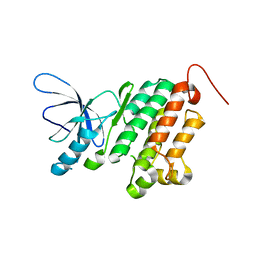 | |
1R8O
 
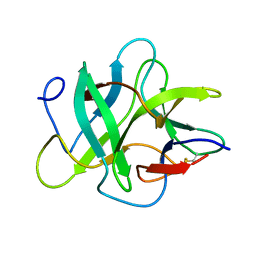 | | Crystal structure of an unusual Kunitz-type trypsin inhibitor from Copaifera langsdorffii seeds | | Descriptor: | Kunitz trypsin inhibitor | | Authors: | Krauchenco, S, Nagem, R.A.P, da Silva, J.A, Marangoni, S, Polikarpov, I. | | Deposit date: | 2003-10-27 | | Release date: | 2004-05-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Three-dimensional structure of an unusual Kunitz (STI) type trypsin inhibitor from Copaifera langsdorffii.
Biochimie, 86, 2004
|
|
3O8M
 
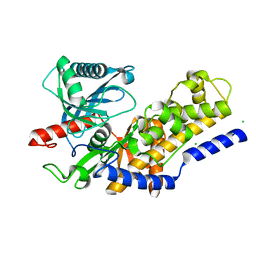 | | Crystal structure of monomeric KlHxk1 in crystal form XI with glucose bound (closed state) | | Descriptor: | CHLORIDE ION, Hexokinase, alpha-D-glucopyranose, ... | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-08-03 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
2V14
 
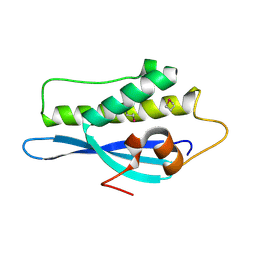 | | Kinesin 16B Phox-homology domain (KIF16B) | | Descriptor: | KINESIN-LIKE MOTOR PROTEIN C20ORF23 | | Authors: | Wilson, M.I, Williams, R.L, Cho, W, Hong, W, Blatner, N.R. | | Deposit date: | 2007-05-21 | | Release date: | 2007-07-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis of Novel Endosome Anchoring Activity of Kif16B Kinesin.
Embo J., 26, 2007
|
|
1RI7
 
 | |
1MNN
 
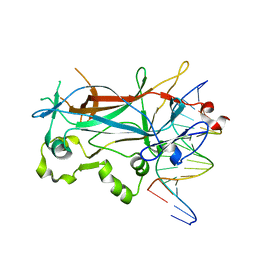 | | Structure of the sporulation specific transcription factor Ndt80 bound to DNA | | Descriptor: | 5'-D(*AP*GP*TP*TP*TP*TP*TP*GP*TP*GP*TP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*AP*CP*AP*CP*AP*AP*AP*AP*AP*C)-3', NDT80 protein | | Authors: | Lamoureux, J.S, Stuart, D, Tsang, R, Wu, C, Glover, J.N. | | Deposit date: | 2002-09-05 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the sporulation-specific transcription factor Ndt80 bound to DNA
Embo J., 21, 2002
|
|
3GC7
 
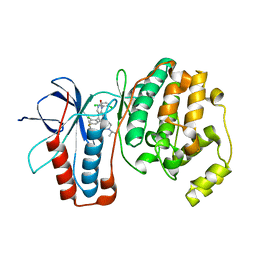 | | The structure of p38alpha in complex with a dihydroquinazolinone | | Descriptor: | 5-(2-chloro-4-fluorophenyl)-1-(2,6-dichlorophenyl)-7-[1-(1-methylethyl)piperidin-4-yl]-3,4-dihydroquinazolin-2(1H)-one, Mitogen-activated protein kinase 14 | | Authors: | Scapin, G, Patel, S.B. | | Deposit date: | 2009-02-21 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of MAP kinase p38beta: different features of the ATP-binding site in p38beta compared with p38alpha.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1VGW
 
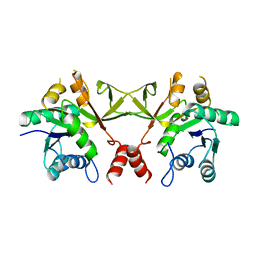 | |
1VHV
 
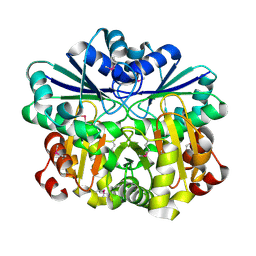 | | Crystal structure of diphthine synthase | | Descriptor: | diphthine synthase | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VIC
 
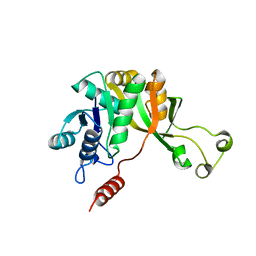 | | Crystal structure of CMP-KDO synthetase | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VIV
 
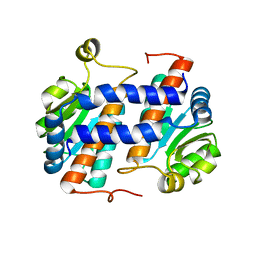 | | Crystal structure of a hypothetical protein | | Descriptor: | Hypothetical protein yckF | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O9H
 
 | |
3BBB
 
 | |
