6AZ0
 
 | | Mitochondrial ATPase Protease YME1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Puchades, C, Rampello, A.J, Shin, M, Giuliano, C, Wiseman, R.L, Glynn, S.E, Lander, G.C. | | Deposit date: | 2017-09-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing.
Science, 358, 2017
|
|
8K53
 
 | |
6BN7
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET23 PROTAC. | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6DFP
 
 | | Crystal Structure of a Tripartite Toxin Component VCA0883 from Vibrio cholerae | | Descriptor: | VCA0883 | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-15 | | Release date: | 2018-05-23 | | Last modified: | 2022-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
8K3J
 
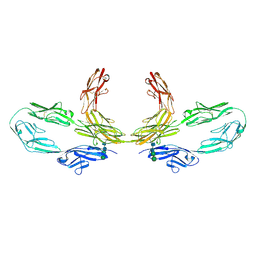 | | Structure of human CNTN2 immunoglobulin domains 1-6 homo-dimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-2, ... | | Authors: | Zhang, Z.Z. | | Deposit date: | 2023-07-16 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human CNTN2 immunoglobulin domains 1-6 homo-dimer
To Be Published
|
|
6BOY
 
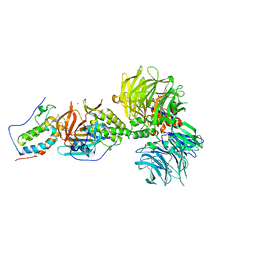 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET6 PROTAC. | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-(8-{[({2-[(3S)-2,6-dioxopiperidin-3-yl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}oxy)acetyl]amino}octyl)acetamide, Bromodomain-containing protein 4, DNA damage-binding protein 1, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-21 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6DWM
 
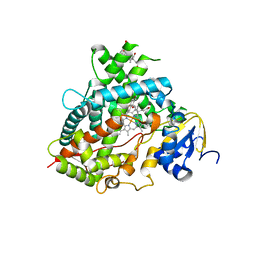 | | Structure of Human Cytochrome P450 1A1 with Bergamottin | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 4-{[(2E)-3,7-dimethylocta-2,6-dien-1-yl]oxy}-7H-furo[3,2-g][1]benzopyran-7-one, Cytochrome P450 1A1, ... | | Authors: | Bart, A.G, Scott, E.E. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of human cytochrome P450 1A1 with bergamottin and erlotinib reveal active-site modifications for binding of diverse ligands.
J. Biol. Chem., 293, 2018
|
|
6BN9
 
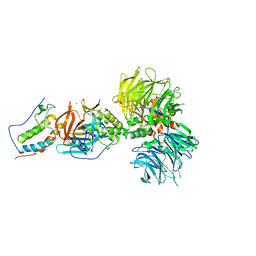 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET70 PROTAC | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.382 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BNB
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET57 PROTAC | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Ishoey, M, He, Z, Zhang, T, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (6.343 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6DWN
 
 | | Structure of Human Cytochrome P450 1A1 with Erlotinib | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Cytochrome P450 1A1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bart, A.G, Scott, E.E. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of human cytochrome P450 1A1 with bergamottin and erlotinib reveal active-site modifications for binding of diverse ligands.
J. Biol. Chem., 293, 2018
|
|
1SOS
 
 | | ATOMIC STRUCTURES OF WILD-TYPE AND THERMOSTABLE MUTANT RECOMBINANT HUMAN CU, ZN SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE, ... | | Authors: | Parge, H.E, Hallewell, R.A, Tainer, J.A. | | Deposit date: | 1992-02-11 | | Release date: | 1993-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic structures of wild-type and thermostable mutant recombinant human Cu,Zn superoxide dismutase.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
5GUH
 
 | | Crystal structure of silkworm PIWI-clade Argonaute Siwi bound to piRNA | | Descriptor: | MAGNESIUM ION, PIWI, RNA (28-MER) | | Authors: | Matsumoto, N, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Silkworm PIWI-Clade Argonaute Siwi Bound to piRNA
Cell, 167, 2016
|
|
4B2D
 
 | | human PKM2 with L-serine and FBP bound. | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, PYRUVATE KINASE ISOZYMES M1/M2, ... | | Authors: | Chaneton, B, Hillmann, P, Zheng, L, Martin, A.C.L, Maddocks, O.D.K, Chokkathukalam, A, Coyle, J.E, Jankevics, A, Holding, F.P, Vousden, K.H, Frezza, C, O'Reilly, M, Gottlieb, E. | | Deposit date: | 2012-07-13 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Serine is a natural ligand and allosteric activator of pyruvate kinase M2.
Nature, 491, 2012
|
|
4RBV
 
 | |
2CCM
 
 | | X-ray structure of Calexcitin from Loligo pealeii at 1.8A | | Descriptor: | CALCIUM ION, CALEXCITIN | | Authors: | Erskine, P.T, Beaven, G.D.E, Wood, S.P, Fox, G, Vernon, J, Giese, K.P, Cooper, J.B. | | Deposit date: | 2006-01-16 | | Release date: | 2006-01-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Neuronal Protein Calexcitin Suggests a Mode of Interaction in Signalling Pathways of Learning and Memory.
J.Mol.Biol., 357, 2006
|
|
3ES5
 
 | | Crystal Structure of Partitivirus (PsV-F) | | Descriptor: | Putative capsid protein | | Authors: | Pan, J, Dong, L, Lin, L, Ochoa, W.F, Sinkovits, R.S, Havens, W.M, Nibert, M.L, Baker, T.S, Ghabrial, S.A, Tao, Y.J. | | Deposit date: | 2008-10-03 | | Release date: | 2009-03-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Atomic structure reveals the unique capsid organization of a dsRNA virus.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2CK3
 
 | | Azide inhibited bovine F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE SUBUNIT ALPHA, MITOCHONDRIAL, ... | | Authors: | Bowler, M.W, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | How Azide Inhibits ATP Hydrolysis by the F-Atpases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
3SCE
 
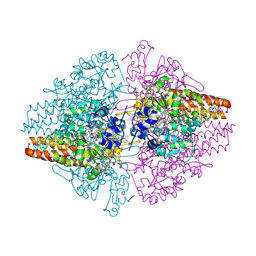 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase with a covalent bond between the CE1 atom of Tyr303 and the CG atom of Gln360 (TvNiRb) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2011-06-07 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1DTK
 
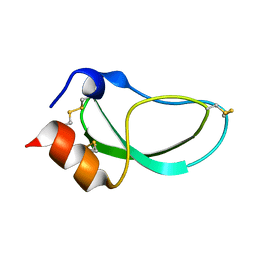 | |
1OJR
 
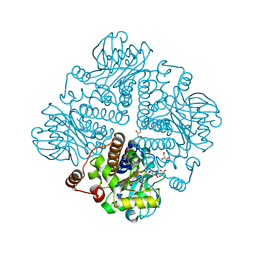 | | L-rhamnulose-1-phosphate aldolase from Escherichia coli (mutant E192A) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Dihydroxyacetone, GLYCEROL, ... | | Authors: | Kroemer, M, Merkel, I, Schulz, G.E. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and Catalytic Mechanism of L-Rhamnulose-1-Phosphate Aldolase.
Biochemistry, 42, 2003
|
|
3Q85
 
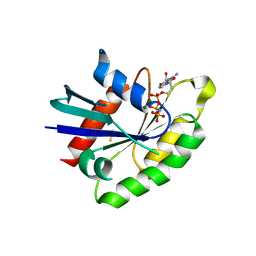 | | Crystal Structure of Rem2 G-domain -GTP Analog Complex | | Descriptor: | GTP-binding protein REM 2, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Navon-Perry, L, Hirsch, J.A. | | Deposit date: | 2011-01-06 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.757 Å) | | Cite: | RGK Family G-Domain:GTP Analog Complex Structures and Nucleotide-Binding Properties.
J.Mol.Biol., 413, 2011
|
|
2N1T
 
 | | Dynamic binding mode of a synaptotagmin-1-SNARE complex in solution | | Descriptor: | Synaptosomal-associated protein 25, Synaptotagmin-1, Syntaxin-1A, ... | | Authors: | Brewer, K, Bacaj, T, Cavalli, A, Camilloni, C, Swarbrick, J, Liu, J, Zhou, A, Zhou, P, Barlow, N, Xu, J, Seven, A, Prinslow, E, Voleti, R, Haussinger, D, Bonvin, A, Tomchick, D, Vendruscolo, M, Graham, B, Sudhof, T, Rizo, J. | | Deposit date: | 2015-04-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dynamic binding mode of a Synaptotagmin-1-SNARE complex in solution.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8QAB
 
 | | X-ray crystal structure of a de novo designed antiparallel coiled-coil hexameric alpha-helical barrel with 4 heptad repeats, apCCHex | | Descriptor: | 1-methoxy-2-[2-[2-[2-[2-[2-[2-[2-[2-[2-(2-methoxyethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethane, apCCHex | | Authors: | Naudin, E.N, Albanese, K.I, Petrenas, R, Woolfson, D.N. | | Deposit date: | 2023-08-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rationally seeded computational protein design of ɑ-helical barrels.
Nat.Chem.Biol., 20, 2024
|
|
8QAD
 
 | |
3MPW
 
 | | Structure of EUTM in 2-D protein membrane | | Descriptor: | Ethanolamine utilization protein eutM, PHOSPHATE ION | | Authors: | Sagermann, M, Takenoya, M, Nikolakakis, K. | | Deposit date: | 2010-04-27 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic insights into the pore structures and mechanisms of the EutL and EutM shell proteins of the ethanolamine-utilizing microcompartment of Escherichia coli.
J.Bacteriol., 192, 2010
|
|
