7YAZ
 
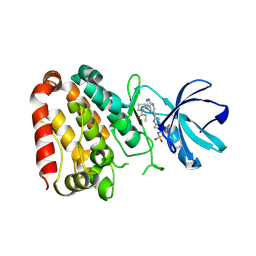 | | Crystal structure of ZAK in complex with compound YH-186 | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, ~{N}-[2,4-bis(fluoranyl)-3-[4-[3-[(3~{S})-1-propanoylpyrrolidin-3-yl]oxy-1~{H}-pyrazolo[3,4-b]pyridin-5-yl]-1,2,3-triazol-1-yl]phenyl]-3-phenyl-benzenesulfonamide | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2022-06-28 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Rational Design of Covalent Kinase Inhibitors by an Integrated Computational Workflow (Kin-Cov).
J.Med.Chem., 66, 2023
|
|
7YAW
 
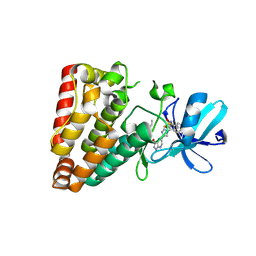 | | Crystal structure of ZAK in complex with compound YH-180 | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, ~{N}-[3-[[5-[1-[2,6-bis(fluoranyl)-3-[(3-phenylphenyl)sulfonylamino]phenyl]-1,2,3-triazol-4-yl]-1~{H}-pyrazolo[3,4-b]pyridin-3-yl]oxy]propyl]propanamide | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2022-06-28 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Design of Covalent Kinase Inhibitors by an Integrated Computational Workflow (Kin-Cov).
J.Med.Chem., 66, 2023
|
|
8XQS
 
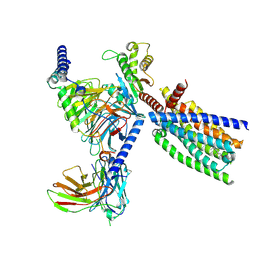 | | Structure of human class T GPCR TAS2R14-DNGi complex with Flufenamic acid. | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
6YII
 
 | |
6SEG
 
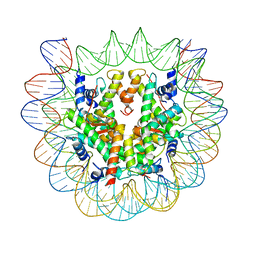 | | Class1: CENP-A nucleosome in complex with CENP-C central region | | Descriptor: | DNA (145-MER), Histone H2A type 2-A, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Ali-Ahmad, A, Bilokapic, S, Schafer, I.B, Halic, M, Sekulic, N. | | Deposit date: | 2019-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail.
Embo Rep., 20, 2019
|
|
7Z7D
 
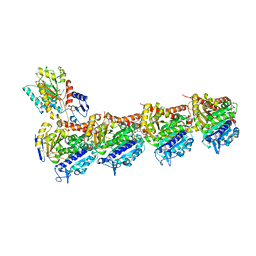 | | Tubulin-Todalam-Vinblastine-complex | | Descriptor: | (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Muehlethaler, T, Milanos, L, Ortega, J.A, Blum, T.B, Gioia, D, Roy, B, Prota, A.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2022-03-15 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational Design of a Novel Tubulin Inhibitor with a Unique Mechanism of Action.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6YMV
 
 | |
1VCC
 
 | |
6SE6
 
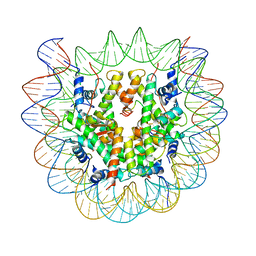 | | Class2 : CENP-A nucleosome in complex with CENP-C central region | | Descriptor: | Centromere protein C, DNA (145-MER), Histone H2A type 2-A, ... | | Authors: | Ali-Ahmad, A, Bilokapic, S, Schafer, I.B, Halic, M, Sekulic, N. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail.
Embo Rep., 20, 2019
|
|
6SO5
 
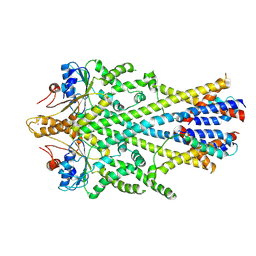 | | Homo sapiens WRB/CAML heterotetramer in complex with a TRC40 dimer | | Descriptor: | ATPase ASNA1, Calcium signal-modulating cyclophilin ligand, Tail-anchored protein insertion receptor WRB, ... | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Flemming, D, Sinning, I. | | Deposit date: | 2019-08-29 | | Release date: | 2020-09-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural Basis of Tail-Anchored Membrane Protein Biogenesis by the GET Insertase Complex.
Mol.Cell, 80, 2020
|
|
6SJF
 
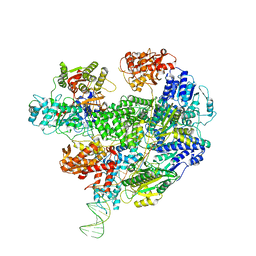 | | Cryo-EM structure of the RecBCD Chi unrecognised complex | | Descriptor: | Forked DNA substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7Y6A
 
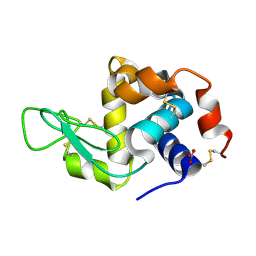 | | Crystal structure of Chicken Egg Lysozyme | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | DeMirci, H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cryogenic X-ray crystallographic studies of biomacromolecules at Turkish Light Source " Turkish DeLight ".
Turk J Biol, 47, 2023
|
|
7XOJ
 
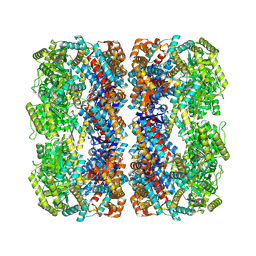 | |
7XON
 
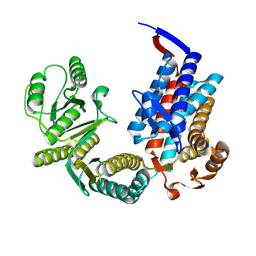 | |
7XYE
 
 | | The apo structure of Orf1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-formimidoyl fortimicin A synthase | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-06-01 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.482 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7XXC
 
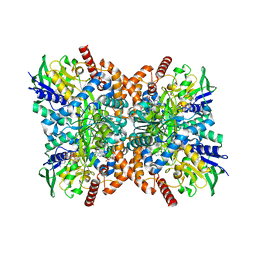 | | Orf1-glycine-glycylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7AEU
 
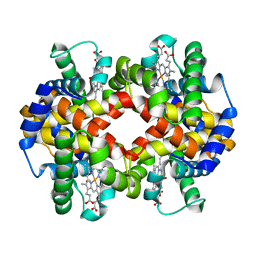 | |
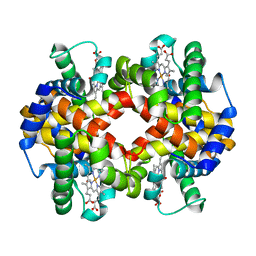
7AEV
 
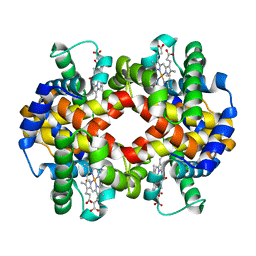 | |
7AET
 
 | |
6SEE
 
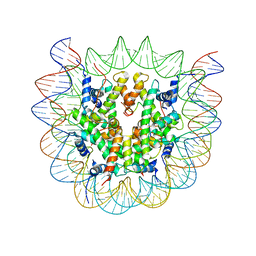 | | Class2A : CENP-A nucleosome in complex with CENP-C central region | | Descriptor: | Centromere protein C, DNA (145-MER), Histone H2A type 2-A, ... | | Authors: | Ali-Ahmad, A, Bilokapic, S, Schafer, I.B, Halic, M, Sekulic, N. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail.
Embo Rep., 20, 2019
|
|
8Y1W
 
 | | Crystal structure of Thermococcus pacificus dUTPase complexed with dUMP. | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Fukui, K, Murakawa, T, Yano, T. | | Deposit date: | 2024-01-25 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | dUTP pyrophosphatases from hyperthermophilic eubacterium and archaeon: Structural and functional examinations on the suitability for PCR application.
Protein Sci., 33, 2024
|
|
6Y8V
 
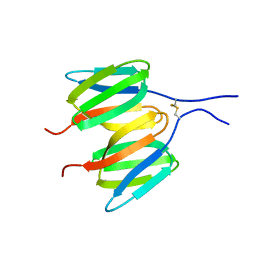 | |
5NF0
 
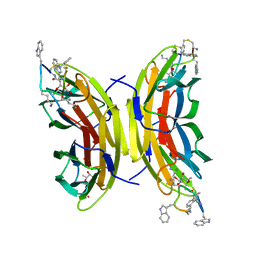 | | Discovery, crystal structures and atomic force microscopy study of thioether ligated D,L-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, CYD-TRP-TRD-LYS-LYD-LYS-LYD-LYS-TRD-TRP-CYD-GLY, ... | | Authors: | Reymond, J.-L, Darbre, T, Stocker, A, Hong, W, van Delden, C, Koehler, T, Luscher, A, Visini, R, Fu, Y, Di Bonaventura, I, He, R. | | Deposit date: | 2017-03-13 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.271 Å) | | Cite: | Design, crystal structure and atomic force microscopy study of thioether ligated d,l-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa.
Chem Sci, 8, 2017
|
|
6Y8W
 
 | |
8Y1Q
 
 | | Crystal structure of Aquifex aeolicus dUTPase complexed with dUMP. | | Descriptor: | 2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Fukui, K, Murakawa, T, Yano, T. | | Deposit date: | 2024-01-25 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | dUTP pyrophosphatases from hyperthermophilic eubacterium and archaeon: Structural and functional examinations on the suitability for PCR application.
Protein Sci., 33, 2024
|
|
