1PDH
 
 | |
2WYQ
 
 | |
1UC8
 
 | | Crystal structure of a lysine biosynthesis enzyme, Lysx, from thermus thermophilus HB8 | | Descriptor: | lysine biosynthesis enzyme | | Authors: | Sakai, H, Vassylyeva, M.N, Matsuura, T, Sekine, S, Nishiyama, M, Terada, T, Shirouzu, M, Kuramitsu, S, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-04-09 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Lysine Biosynthesis Enzyme, LysX, from Thermus thermophilus HB8
J.Mol.Biol., 332, 2003
|
|
3NBQ
 
 | |
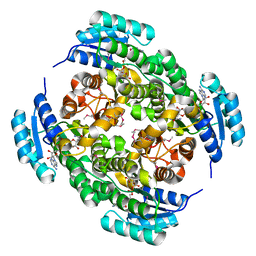
3NDR
 
 | |
1PBC
 
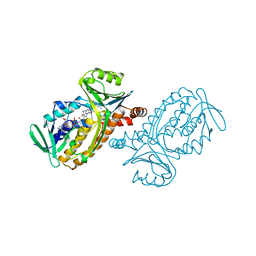 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2-HYDROXY-4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Van Der Bolt, F.J.T, Van Berkel, W.J.H. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
3ETL
 
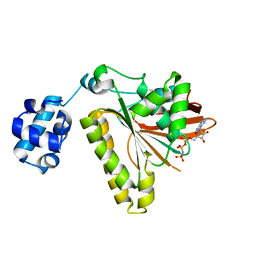 | | RadA recombinase from Methanococcus maripaludis in complex with AMPPNP | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Li, Y, He, Y, Luo, Y. | | Deposit date: | 2008-10-08 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conservation of a conformational switch in RadA recombinase from Methanococcus maripaludis.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2Z8F
 
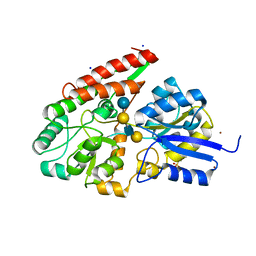 | | The galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum in complex with lacto-N-tetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Galacto-N-biose/lacto-N-biose I transporter substrate-binding protein, SODIUM ION, ... | | Authors: | Suzuki, R, Wada, J, Katayama, T, Fushinobu, S. | | Deposit date: | 2007-09-05 | | Release date: | 2008-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I.
J.Biol.Chem., 283, 2008
|
|
1PIW
 
 | | APO AND HOLO STRUCTURES OF AN NADP(H)-DEPENDENT CINNAMYL ALCOHOL DEHYDROGENASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | Hypothetical zinc-type alcohol dehydrogenase-like protein in PRE5-FET4 intergenic region, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Valencia, E, Larroy, C, Ochoa, W.F, Pares, X, Fita, I, Biosca, J.A. | | Deposit date: | 2003-05-30 | | Release date: | 2004-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Apo and Holo Structures of an NADP(H)-dependent Cinnamyl Alcohol Dehydrogenase from Saccharomyces cerevisiae
J.Mol.Biol., 341, 2004
|
|
1UB8
 
 | | Crystal structure of d(GCGAAGC), bending duplex with a bulge-in residue | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*GP*C)-3', COBALT HEXAMMINE(III) | | Authors: | Sunami, T, Kondo, J, Hirao, I, Watanabe, K, Miura, K, Takenaka, A. | | Deposit date: | 2003-03-31 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of d(GCGAAGC) and d(GCGAAAGC) (tetragonal form): a switching of partners of the sheared G.A pairs to form a functional G.AxA.G crossing.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3EKJ
 
 | | Calcium-free GCaMP2 (calcium binding deficient mutant) | | Descriptor: | Myosin light chain kinase, Green fluorescent protein, Calmodulin chimera | | Authors: | Akerboom, J, Velez Rivera, J.D, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2008-09-19 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of the GCaMP Calcium Sensor Reveal the Mechanism of Fluorescence Signal Change and Aid Rational Design
J.Biol.Chem., 284, 2009
|
|
1K9Q
 
 | | YAP65 WW domain complexed to N-(n-octyl)-GPPPY-NH2 | | Descriptor: | 65 kDa Yes-associated protein, N-OCTANE, WW domain binding protein-1 | | Authors: | Pires, J.R, Taha-Nejad, F, Toepert, F, Ast, T, Hoffmuller, U, Schneider-Mergener, J, Kuhne, R, Macias, M.J, Oschkinat, H. | | Deposit date: | 2001-10-30 | | Release date: | 2001-12-28 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the YAP65 WW domain and the variant L30 K in complex with the peptides GTPPPPYTVG, N-(n-octyl)-GPPPY and PLPPY and the application of peptide libraries reveal a minimal binding epitope.
J.Mol.Biol., 314, 2001
|
|
1K9W
 
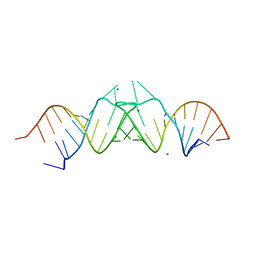 | | HIV-1(MAL) RNA Dimerization Initiation Site | | Descriptor: | HIV-1 DIS(Mal)UU RNA, MAGNESIUM ION | | Authors: | Ennifar, E, Walter, P, Ehresmann, B, Ehresmann, C, Dumas, P. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site.
Nat.Struct.Biol., 8, 2001
|
|
1PMY
 
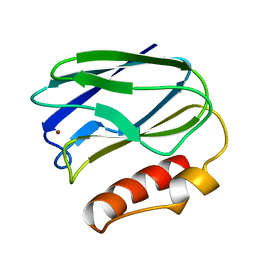 | | REFINED CRYSTAL STRUCTURE OF PSEUDOAZURIN FROM METHYLOBACTERIUM EXTORQUENS AM1 AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Kai, Y, Harada, S, Kasai, N, Ohshiro, Y, Suzuki, S, Kohzuma, T, Tobari, J. | | Deposit date: | 1994-01-28 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Refined crystal structure of pseudoazurin from Methylobacterium extorquens AM1 at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
3O0G
 
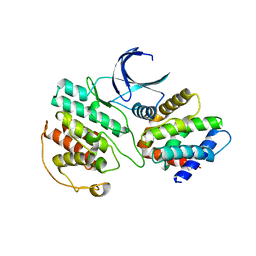 | | Crystal Structure of Cdk5:p25 in complex with an ATP analogue | | Descriptor: | Cell division protein kinase 5, Cyclin-dependent kinase 5 activator 1, {4-amino-2-[(4-chlorophenyl)amino]-1,3-thiazol-5-yl}(3-nitrophenyl)methanone | | Authors: | Mapelli, M. | | Deposit date: | 2010-07-19 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Defining Cdk5 ligand chemical space with small molecule inhibitors of Tau phosphorylation
Chem.Biol., 12, 2005
|
|
1PSK
 
 | |
1PSZ
 
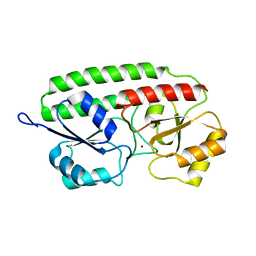 | | PNEUMOCOCCAL SURFACE ANTIGEN PSAA | | Descriptor: | PROTEIN (SURFACE ANTIGEN PSAA), ZINC ION | | Authors: | Lawrence, M.C, Pilling, P.A, Epa, V.C, Berry, A.M, Ogunniyi, A.D, Paton, J.C. | | Deposit date: | 1998-10-13 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of pneumococcal surface antigen PsaA reveals a metal-binding site and a novel structure for a putative ABC-type binding protein.
Structure, 6, 1998
|
|
1PO2
 
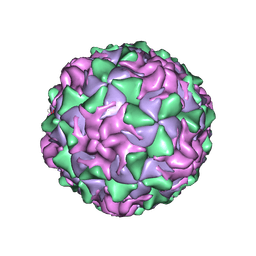 | | POLIOVIRUS (TYPE 1, MAHONEY) IN COMPLEX WITH R77975, AN INHIBITOR OF VIRAL REPLICATION | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE ETHYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 1 MAHONEY | | Authors: | Hiremath, C.N, Filman, D.J, Grant, R.A, Hogle, J.M. | | Deposit date: | 1997-01-08 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand-induced conformational changes in poliovirus-antiviral drug complexes.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
2X3A
 
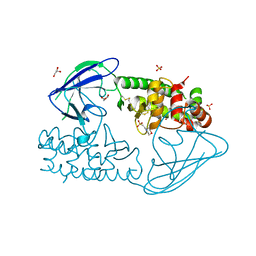 | | AsaP1 inactive mutant E294Q, an extracellular toxic zinc metalloendopeptidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Bogdanovic, X, Palm, G.J, Singh, R.K, Hinrichs, W. | | Deposit date: | 2010-01-22 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Evidence of Intramolecular Propeptide Inhibition of the Aspzincin Metalloendopeptidase Asap1.
FEBS Lett., 590, 2016
|
|
1K9R
 
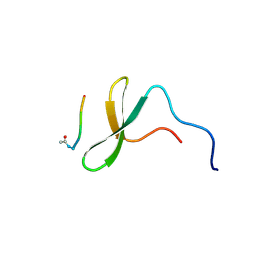 | | YAP65 WW domain complexed to Acetyl-PLPPY | | Descriptor: | 65 kDa Yes-associated protein, WW domain binding protein-1 | | Authors: | Pires, J.R, Taha-Nejad, F, Toepert, F, Ast, T, Hoffmuller, U, Schneider-Mergener, J, Kuhne, R, Macias, M.J, Oschkinat, H. | | Deposit date: | 2001-10-30 | | Release date: | 2001-12-28 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the YAP65 WW domain and the variant L30 K in complex with the peptides GTPPPPYTVG, N-(n-octyl)-GPPPY and PLPPY and the application of peptide libraries reveal a minimal binding epitope.
J.Mol.Biol., 314, 2001
|
|
1PQJ
 
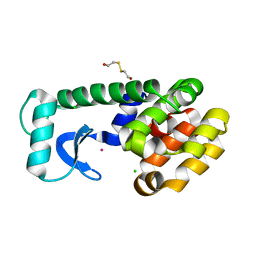 | | T4 LYSOZYME CORE REPACKING MUTANT A111V/CORE10/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Lysozyme, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-06-18 | | Release date: | 2003-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | REPACKING THE CORE OF T4 LYSOZYME BY AUTOMATED DESIGN
J.Mol.Biol., 332, 2003
|
|
1K40
 
 | |
3EQS
 
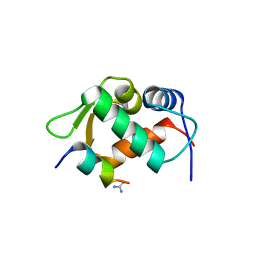 | |
1PS1
 
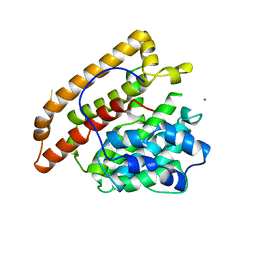 | | PENTALENENE SYNTHASE | | Descriptor: | PENTALENENE SYNTHASE, TRIMETHYL LEAD ION | | Authors: | Lesburg, C.A, Christianson, D.W. | | Deposit date: | 1997-03-23 | | Release date: | 1998-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology.
Science, 277, 1997
|
|
1PSR
 
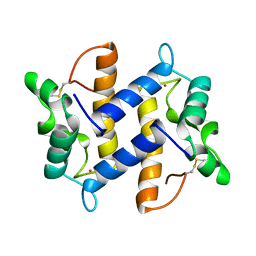 | | HUMAN PSORIASIN (S100A7) | | Descriptor: | HOLMIUM ATOM, PSORIASIN | | Authors: | Brodersen, D.E, Etzerodt, M, Madsen, P, Celis, J, Thoegersen, H.C, Nyborg, J, Kjeldgaard, M. | | Deposit date: | 1997-11-27 | | Release date: | 1999-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | EF-hands at atomic resolution: the structure of human psoriasin (S100A7) solved by MAD phasing.
Structure, 6, 1998
|
|
