9CHR
 
 | |
9FJD
 
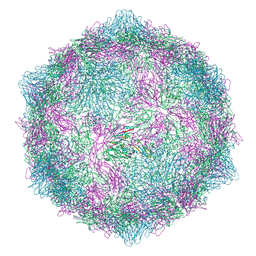 | | Expanded CVB1-VLP (Tween80) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Plavec, Z, Butcher, S.J. | | Deposit date: | 2024-05-31 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Comparison of structure and immunogenicity of CVB1-VLP and inactivated CVB1 vaccine candidates.
Res Sq, 2024
|
|
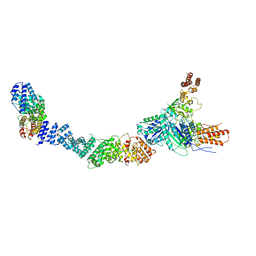
9FA4
 
 | |
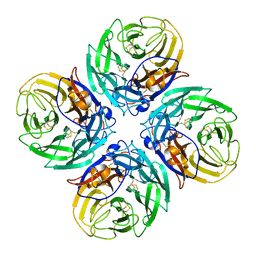
9EWQ
 
 | |
8Y86
 
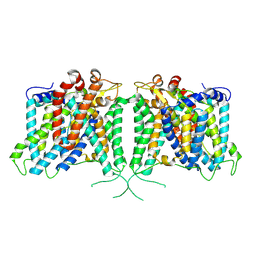 | | Human AE3 with NaHCO3- | | Descriptor: | Anion exchange protein 3, BICARBONATE ION | | Authors: | Jian, L, Zhang, Q, Yao, D, Cao, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | The structural insight into the functional modulation of human anion exchanger 3.
Nat Commun, 15, 2024
|
|
9B9V
 
 | |
9EWX
 
 | | Cryo-EM structure of the Pseudomonas aeruginosa PAO1 Type IV pilus | | Descriptor: | Pilin | | Authors: | Ochner, H, Boehning, J, Wang, Z, Tarafder, A, Caspy, I, Bharat, T.A.M. | | Deposit date: | 2024-04-05 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structure of the Pseudomonas aeruginosa PAO1 Type-IV pilus
To Be Published
|
|
8XZ3
 
 | |
8YBJ
 
 | | Cryo-EM structure of human nucleosome core particle composed of the Widom 601 DNA sequence | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kimura, T, Hirai, S, Kujirai, T, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structure and biochemical analyses of the nucleosome containing the cancer-associated histone H3 mutation E97K.
Genes Cells, 29, 2024
|
|
9C57
 
 | | Reconstituted P400 Subcomplex of the human TIP60 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Yang, Z, Mameri, A, Florez Ariza, A.J, Cote, J, Nogales, E. | | Deposit date: | 2024-06-05 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural insights into the human NuA4/TIP60 acetyltransferase and chromatin remodeling complex.
Science, 385, 2024
|
|
9FFG
 
 | | Empty capsid of Rhodobacter microvirus Ebor computed with I4 symmetry | | Descriptor: | Major capsid protein | | Authors: | Bardy, P, MacDonald, C.I.W, Jenkins, H.T, Byrom, L, Chechik, M, Hart, S.J, Turkenburg, J.P, Blaza, J.N, Fogg, P.C.M, Antson, A.A. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A stargate mechanism of Microviridae genome delivery unveiled by cryogenic electron tomography.
Biorxiv, 2024
|
|
9EO6
 
 | | SARS-CoV2 major protease in complex with a covalent inhibitor SLL11. | | Descriptor: | 3C-like proteinase nsp5, Inhibitor SLL11, POTASSIUM ION | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-14 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 2024
|
|
9BEW
 
 | |
8ZHH
 
 | | SARS-CoV-2 spike trimer (6P) in complex with two H18 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (5.55 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein helping to drive the genesis of Omicron variants
To Be Published
|
|
9F5S
 
 | | EVA71 E096A native particle | | Descriptor: | SPHINGOSINE, VP0, VP1, ... | | Authors: | Kingston, N.J, Stonehouse, N.J, Rowlands, D.J, Hogle, J.M, Filman, D.J, Snowden, J.S.S. | | Deposit date: | 2024-04-30 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Reappraisal of the mechanism of the enterovirus VP0 maturation cleavage based on the structure of a stabilised assembly intermediate
To Be Published
|
|
9FP6
 
 | |
9EMA
 
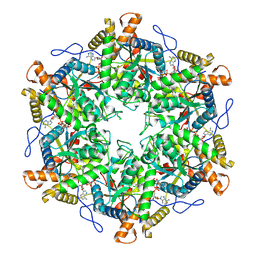 | | RUVBL1/2 in complex with ATP and CB-6644 inhibitor | | Descriptor: | 5-chloranyl-2-ethoxy-4-fluoranyl-~{N}-[4-[[3-(methoxymethyl)-1-oxidanylidene-6,7-dihydro-5~{H}-pyrazolo[1,2-a][1,2]benzodiazepin-2-yl]amino]-2,2-dimethyl-4-oxidanylidene-butyl]benzamide, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lopez-Perrote, A, Llorca, O, Garcia-Martin, C. | | Deposit date: | 2024-03-07 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanism of allosteric inhibition of RUVBL1-RUVBL2 by the small-molecule CB-6644
Cell Rep Phys Sci, 2024
|
|
8YXM
 
 | |
8ZH8
 
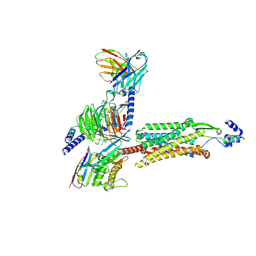 | | Human GPR103 -Gq complex bound to QRFP26 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2,Guanine nucleotide-binding protein G(i) subunit alpha-2,Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Iwama, A, Akasaka, H, Sano, F.K, Oshima, H.S, Shihoya, W, Nureki, O. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure and dynamics of the pyroglutamylated RF-amide peptide QRFP receptor GPR103.
Nat Commun, 15, 2024
|
|
8Y8C
 
 | | Structure of HCoV-HKU1C spike in the inactive-closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8YWX
 
 | |
8ZHE
 
 | | SARS-CoV-2 spike trimer (6P) in complex with three R1-26 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-26 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein helping to drive the genesis of Omicron variants
To Be Published
|
|
9ESM
 
 | |
8YRO
 
 | | SARS-CoV-2 Delta Spike in complex with JL-8C | | Descriptor: | JL-8C Heavy Chain, JL-8C Light Chain, Spike glycoprotein | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2024-03-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8ZHN
 
 | | SARS-CoV-2 spike trimer (6P) in complex with three H18 and three R1-32 Fabs (one RBD rotated) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein helping to drive the genesis of Omicron variants
To Be Published
|
|
