8BHQ
 
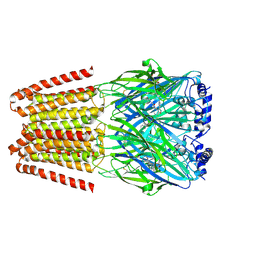 | | GABA-A receptor a5 homomer - a5V3 - RO7172670 | | Descriptor: | 2-[[5-methyl-3-(6-methylpyridazin-3-yl)-1,2-oxazol-4-yl]methyl]-5-(5-oxa-2-azaspiro[3.5]nonan-2-yl)pyridazin-3-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Kasaragod, V.B. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHS
 
 | | GABA-A receptor a5 homomer - a5V3 - RO4938581 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[bis(fluoranyl)methyl]-15-bromanyl-2,4,8,9,11-pentazatetracyclo[11.4.0.0^{2,6}.0^{8,12}]heptadeca-1(13),3,5,9,11,14,16-heptaene, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHA
 
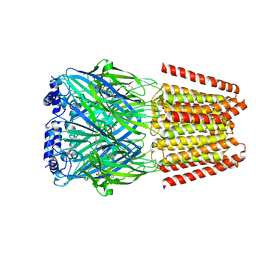 | | GABA-A receptor a5 homomer - a5V3 - Basmisanil - HR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Basmisanil, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Malinauskas, T.M, Wahid, A.A, Hardwick, S.W, Chirgadze, D.Y, Miller, P.S. | | Deposit date: | 2022-10-30 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHR
 
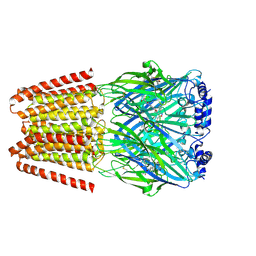 | | GABA-A receptor a5 homomer - a5V3 - RO7015738 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[[5-methyl-3-(6-methylpyridin-3-yl)-1,2-oxazol-4-yl]methoxy]-~{N}-[(2~{S})-1-oxidanylpentan-2-yl]pyridine-3-carboxamide, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Miller, P.S, Malinauskas, T.M, Kasaragod, V.B. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHB
 
 | | GABA-A receptor a5 homomer - a5V3 - RO154513 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, ethyl 8-[(azanylidene-$l^{4}-azanylidene)amino]-5-methyl-6-oxidanylidene-4~{H}-imidazo[1,5-a][1,4]benzodiazepine-3-carboxylate | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y, Wahid, A.A. | | Deposit date: | 2022-10-30 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHG
 
 | | GABA-A receptor a5 heteromer - a5V2 - Bretazenil | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bretazenil, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Miller, P.S, Malinauskas, T.M, Omari, K.E, Aricescu, A.R. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BGI
 
 | | GABA-A receptor a5 homomer - a5V1 - Flumazenil | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, Pregnanolone, ... | | Authors: | Miller, P.S, Malinauskas, T.M, Omari, K.E, Aricescu, A.R. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2IIF
 
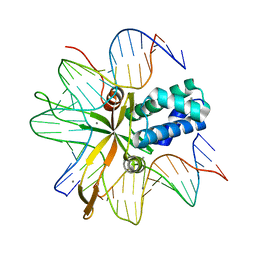 | | single chain Integration Host Factor mutant protein (scIHF2-K45aE) in complex with DNA | | Descriptor: | DNA (5'-D(*DGP*DCP*DTP*DTP*DAP*DTP*DCP*DAP*DAP*DTP*DTP*DTP*DGP*DTP*DTP*DGP*DCP*DAP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DCP*DAP*DAP*DAP*DAP*DAP*DAP*DGP*DCP*DAP*DTP*DT)-3'), Integration host factor, ... | | Authors: | Bao, Q, Droege, P, Davey, C.A. | | Deposit date: | 2006-09-28 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A Divalent Metal-mediated Switch Controlling Protein-induced DNA Bending
J.Mol.Biol., 367, 2007
|
|
6P1U
 
 | | Post-catalytic nicked complex of human DNA Polymerase Mu with 1-nt gapped substrate containing template 8OG and newly incorporated CMP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
7ADH
 
 | |
2IN9
 
 | | crystal structure of Mtu recA intein, splicing domain | | Descriptor: | Endonuclease PI-MtuI, SULFATE ION | | Authors: | Van Roey, P. | | Deposit date: | 2006-10-06 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic and mutational studies of Mycobacterium tuberculosis recA mini-inteins suggest a pivotal role for a highly conserved aspartate residue.
J.Mol.Biol., 367, 2007
|
|
6P8I
 
 | | N-terminal 5 domains of IGFIIR | | Descriptor: | Cation-independent mannose-6-phosphate receptor, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Olson, L.J, Dahms, N.M, Kim, J.-J.P. | | Deposit date: | 2019-06-07 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol, 3, 2020
|
|
3I11
 
 | |
3I0V
 
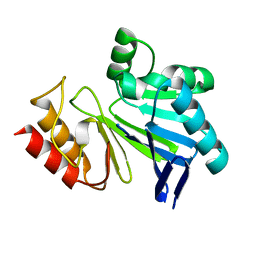 | |
6RRG
 
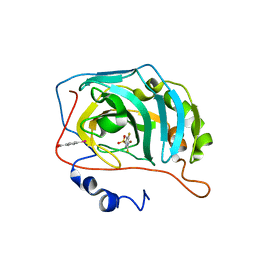 | | Human Carbonic Anhydrase II in complex with fluorinated benzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 3,5-DIFLUOROBENZENESULFONAMIDE, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-05-17 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.127 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
6RS5
 
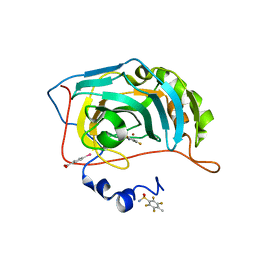 | | Human Carbonic Anhydrase II in complex with fluorinated benzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 2,3,5,6-tetrakis(fluoranyl)-4-methyl-benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Ngo, K, Heine, A, Klebe, G. | | Deposit date: | 2019-05-21 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
1GS4
 
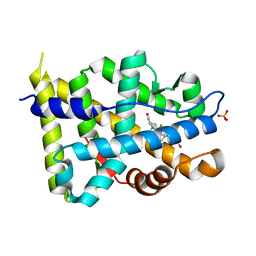 | | Structural basis for the glucocorticoid response in a mutant human androgen receptor (ARccr) derived from an androgen-independent prostate cancer | | Descriptor: | 9ALPHA-FLUOROCORTISOL, ANDROGEN RECEPTOR, PHOSPHATE ION | | Authors: | Matias, P.M, Carrondo, M.A, Coelho, R, Thomaz, M, Zhao, X.-Y, Wegg, A, Crusius, K, Egner, U, Donner, P. | | Deposit date: | 2001-12-27 | | Release date: | 2003-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for the Glucocorticoid Response in a Mutant Human Androgen Receptor (Ar(Ccr)) Derived from an Androgen-Independent Prostate Cancer
J.Med.Chem., 45, 2002
|
|
1UFU
 
 | | Crystal structure of ligand binding domain of immunoglobulin-like transcript 2 (ILT2; LIR-1) | | Descriptor: | Immunoglobulin-like transcript 2 | | Authors: | Shiroishi, M, Amano, K, Rasubala, L, Tsumoto, K, Kumagai, I, Kohda, D, Maenaka, K. | | Deposit date: | 2003-06-10 | | Release date: | 2004-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Kinetic and thermodynamic properties of the interaction between Immunoglobulin like transcript (ILT) and MHC class I
To be Published
|
|
6FZJ
 
 | | PPAR gamma mutant complex | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, CHLORIDE ION, IODIDE ION, ... | | Authors: | Rochel, N. | | Deposit date: | 2018-03-14 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Recurrent activating mutations of PPAR gamma associated with luminal bladder tumors.
Nat Commun, 10, 2019
|
|
1R2T
 
 | |
3L6C
 
 | | X-ray crystal structure of rat serine racemase in complex with malonate a potent inhibitor | | Descriptor: | MALONATE ION, MANGANESE (II) ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Smith, M.A, Mack, V, Ebneth, A, Moraes, I, Felicetti, B, Wood, M, Schonfeld, D, Mather, O, Cesura, A, Barker, J. | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
J.Biol.Chem., 285, 2010
|
|
2EFN
 
 | | Crystal Structure of Ser 32 to Ala of ST1022 from Sulfolobus tokodaii 7 | | Descriptor: | 150aa long hypothetical transcriptional regulator, MAGNESIUM ION | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-23 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding.
Nucleic Acids Res., 36, 2008
|
|
6MIU
 
 | |
3I15
 
 | |
9CXA
 
 | | Native human GABAA receptor of beta2-alpha1-beta3-alpha1-gamma2 assembly | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, J, Hibbs, R.E, Noviello, C.M. | | Deposit date: | 2024-07-31 | | Release date: | 2025-01-22 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Resolving native GABA A receptor structures from the human brain.
Nature, 638, 2025
|
|
