1BVD
 
 | | STRUCTURE OF A BILIVERDIN APOMYOGLOBIN COMPLEX (FORM B) AT 98 K | | Descriptor: | APOMYOGLOBIN, BILIVERDINE IX ALPHA | | Authors: | Wagner, U.G, Mueller, N, Schmitzberger, W, Falk, H, Kratky, C. | | Deposit date: | 1994-12-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure determination of the biliverdin apomyoglobin complex: crystal structure analysis of two crystal forms at 1.4 and 1.5 A resolution.
J.Mol.Biol., 247, 1995
|
|
1BVE
 
 | | HIV-1 PROTEASE-DMP323 COMPLEX IN SOLUTION, NMR, 28 STRUCTURES | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Yamazaki, T, Hinck, A.P, Wang, Y.-X, Nicholson, L.K, Torchia, D.A, Wingfield, P, Stahl, S.J, Kaufman, J.D, Chang, C, Domaille, P.J, Lam, P.Y.S. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the HIV-1 protease complexed with DMP323, a novel cyclic urea-type inhibitor, determined by nuclear magnetic resonance spectroscopy.
Protein Sci., 5, 1996
|
|
1BVG
 
 | | HIV-1 PROTEASE-DMP323 COMPLEX IN SOLUTION, NMR MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Yamazaki, T, Hinck, A.P, Wang, Y.-X, Nicholson, L.K, Torchia, D.A, Wingfield, P, Stahl, S.J, Kaufman, J.D, Chang, C, Domaille, P.J, Lam, P.Y.S. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the HIV-1 protease complexed with DMP323, a novel cyclic urea-type inhibitor, determined by nuclear magnetic resonance spectroscopy.
Protein Sci., 5, 1996
|
|
1BVH
 
 | | SOLUTION STRUCTURE OF A LOW MOLECULAR WEIGHT PROTEIN TYROSINE PHOSPHATASE | | Descriptor: | ACID PHOSPHATASE | | Authors: | Logan, T.M, Zhou, M.-M, Nettesheim, D.G, Meadows, R.P, Van Etten, R.L, Fesik, S.W. | | Deposit date: | 1994-05-03 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a low molecular weight protein tyrosine phosphatase.
Biochemistry, 33, 1994
|
|
1BVI
 
 | | RIBONUCLEASE T1 (WILDTYPE) COMPLEXED WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Langhorst, U, Loris, R, Denisov, V.P, Doumen, J, Roose, P, Maes, D, Halle, B, Steyaert, J. | | Deposit date: | 1998-09-15 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissection of the structural and functional role of a conserved hydration site in RNase T1.
Protein Sci., 8, 1999
|
|
1BVJ
 
 | | HIV-1 RNA A-RICH HAIRPIN LOOP | | Descriptor: | RNA (5'-R(P*GP*GP*CP*GP*AP*CP*GP*GP*UP*GP*UP*AP*AP*AP*AP*AP*UP*CP*UP*CP*GP*CP* C)-3') | | Authors: | Puglisi, E.V, Puglisi, J.D. | | Deposit date: | 1998-08-31 | | Release date: | 1998-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HIV-1 A-rich RNA loop mimics the tRNA anticodon structure.
Nat.Struct.Biol., 5, 1998
|
|
1BVK
 
 | |
1BVL
 
 | | HUMANIZED ANTI-LYSOZYME FV | | Descriptor: | HULYS11 | | Authors: | Holmes, M.A, Foote, J. | | Deposit date: | 1998-09-16 | | Release date: | 1999-02-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural consequences of humanizing an antibody.
J.Immunol., 158, 1997
|
|
1BVM
 
 | | SOLUTION NMR STRUCTURE OF BOVINE PANCREATIC PHOSPHOLIPASE A2, 20 STRUCTURES | | Descriptor: | PROTEIN (PHOSPHOLIPASE A2) | | Authors: | Yuan, C.-H, Byeon, I.-J.L, Li, Y, Tsai, M.-D. | | Deposit date: | 1998-09-14 | | Release date: | 1999-09-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of phospholipase A2 from functional perspective. 1. Functionally relevant solution structure and roles of the hydrogen-bonding network.
Biochemistry, 38, 1999
|
|
1BVN
 
 | | PIG PANCREATIC ALPHA-AMYLASE IN COMPLEX WITH THE PROTEINACEOUS INHIBITOR TENDAMISTAT | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROTEIN (ALPHA-AMYLASE), ... | | Authors: | Machius, M, Wiegand, G, Epp, O, Huber, R. | | Deposit date: | 1998-09-16 | | Release date: | 1998-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of porcine pancreatic alpha-amylase in complex with the microbial inhibitor Tendamistat.
J.Mol.Biol., 247, 1995
|
|
1BVO
 
 | | DORSAL HOMOLOGUE GAMBIF1 BOUND TO DNA | | Descriptor: | DNA DUPLEX, TRANSCRIPTION FACTOR GAMBIF1 | | Authors: | Cramer, P, Varrot, A, Barillas-Mury, C, Kafatos, F.C, Mueller, C.W. | | Deposit date: | 1998-09-16 | | Release date: | 1999-07-12 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the specificity domain of the Dorsal homologue Gambif1 bound to DNA.
Structure Fold.Des., 7, 1999
|
|
1BVP
 
 | |
1BVQ
 
 | |
1BVR
 
 | | M.TB. ENOYL-ACP REDUCTASE (INHA) IN COMPLEX WITH NAD+ AND C16-FATTY-ACYL-SUBSTRATE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (ENOYL-ACYL CARRIER PROTEIN (ACP) REDUCTASE), TRANS-2-HEXADECENOYL-(N-ACETYL-CYSTEAMINE)-THIOESTER | | Authors: | Rozwarski, D.A, Vilcheze, C, Sugantino, M, Bittman, R, Jacobs, W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1998-09-17 | | Release date: | 1999-09-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis enoyl-ACP reductase, InhA, in complex with NAD+ and a C16 fatty acyl substrate.
J.Biol.Chem., 274, 1999
|
|
1BVT
 
 | | METALLO-BETA-LACTAMASE FROM BACILLUS CEREUS 569/H/9 | | Descriptor: | BICARBONATE ION, PROTEIN (BETA-LACTAMASE), ZINC ION | | Authors: | Carfi, A, Duee, E, Dideberg, O. | | Deposit date: | 1998-09-18 | | Release date: | 1998-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 A resolution structure of the zinc (II) beta-lactamase from Bacillus cereus.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1BVU
 
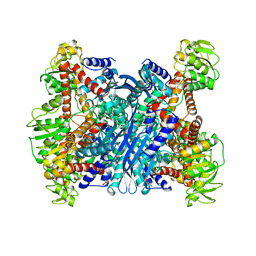 | | GLUTAMATE DEHYDROGENASE FROM THERMOCOCCUS LITORALIS | | Descriptor: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | Authors: | Baker, P.J, Britton, K.L, Yip, K.S, Stillman, T.J, Rice, D.W. | | Deposit date: | 1999-07-20 | | Release date: | 1999-09-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure determination of the glutamate dehydrogenase from the hyperthermophile Thermococcus litoralis and its comparison with that from Pyrococcus furiosus
J.Mol.Biol., 293, 1999
|
|
1BVV
 
 | |
1BVW
 
 | | CELLOBIOHYDROLASE II (CEL6A) FROM HUMICOLA INSOLENS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Varrot, A, Davies, G.J, Schulein, M. | | Deposit date: | 1998-09-19 | | Release date: | 1999-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of the catalytic core domain of the family 6 cellobiohydrolase II, Cel6A, from Humicola insolens, at 1.92 A resolution.
Biochem.J., 337, 1999
|
|
1BVX
 
 | | THE 1.8 A STRUCTURE OF GEL GROWN TETRAGONAL HEN EGG WHITE LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME) | | Authors: | Dong, J, Boggon, T.J, Chayen, N.E, Raftery, J, Bi, R.C, Helliwell, J.R. | | Deposit date: | 1998-09-18 | | Release date: | 1998-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bound-solvent structures for microgravity-, ground control-, gel- and microbatch-grown hen egg-white lysozyme crystals at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BVY
 
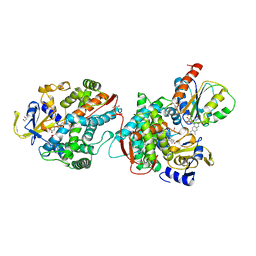 | | COMPLEX OF THE HEME AND FMN-BINDING DOMAINS OF THE CYTOCHROME P450(BM-3) | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, PROTEIN (CYTOCHROME P450 BM-3), ... | | Authors: | Sevrioukova, I.F, Li, H, Zhang, H, Peterson, J.A, Poulos, T.L. | | Deposit date: | 1998-09-21 | | Release date: | 1999-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of a cytochrome P450-redox partner electron-transfer complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1BVZ
 
 | | ALPHA-AMYLASE II (TVAII) FROM THERMOACTINOMYCES VULGARIS R-47 | | Descriptor: | PROTEIN (ALPHA-AMYLASE II) | | Authors: | Kamitori, S, Kondo, S, Okuyama, K, Yokota, T, Shimura, Y, Tonozuka, T, Sakano, Y. | | Deposit date: | 1998-09-22 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase II (TVAII) hydrolyzing cyclodextrins and pullulan at 2.6 A resolution.
J.Mol.Biol., 287, 1999
|
|
1BW0
 
 | | CRYSTAL STRUCTURE OF TYROSINE AMINOTRANSFERASE FROM TRYPANOSOMA CRUZI | | Descriptor: | PROTEIN (TYROSINE AMINOTRANSFERASE) | | Authors: | Blankenfeldt, W, Montemartini, M, Hunter, G.R, Kalisz, H.M, Nowicki, C, Hecht, H.J. | | Deposit date: | 1998-09-28 | | Release date: | 1999-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Trypanosoma cruzi tyrosine aminotransferase: substrate specificity is influenced by cofactor binding mode.
Protein Sci., 8, 1999
|
|
1BW3
 
 | |
1BW4
 
 | |
1BW5
 
 | | THE NMR SOLUTION STRUCTURE OF THE HOMEODOMAIN OF THE RAT INSULIN GENE ENHANCER PROTEIN ISL-1, 50 STRUCTURES | | Descriptor: | INSULIN GENE ENHANCER PROTEIN ISL-1 | | Authors: | Ippel, J.H, Larsson, G, Behravan, G, Zdunek, J, Lundqvist, M, Schleucher, J, Lycksell, P.-O, Wijmenga, S.S. | | Deposit date: | 1998-09-29 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the homeodomain of the rat insulin-gene enhancer protein isl-1. Comparison with other homeodomains.
J.Mol.Biol., 288, 1999
|
|
