4X33
 
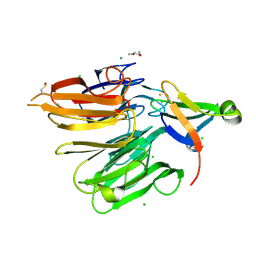 | | Structure of the Elongator cofactor complex Kti11/Kti13 at 1.45A | | Descriptor: | 1,2-DIMETHOXYETHANE, CHLORIDE ION, Diphthamide biosynthesis protein 3, ... | | Authors: | Kolaj-Robin, O, McEwen, A.G, Cavarelli, J, Seraphin, B. | | Deposit date: | 2014-11-27 | | Release date: | 2015-01-21 | | Last modified: | 2015-03-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the Elongator cofactor complex Kti11/Kti13 provides insight into the role of Kti13 in Elongator-dependent tRNA modification.
Febs J., 282, 2015
|
|
5TED
 
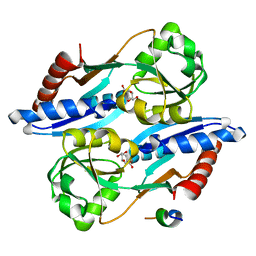 | | Effector binding domain of QuiR in complex with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, His Tag peptide, Lmo0488 protein | | Authors: | Prezioso, S.M, Christendat, D. | | Deposit date: | 2016-09-21 | | Release date: | 2017-10-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Shikimate Induced Transcriptional Activation of Protocatechuate Biosynthesis Genes by QuiR, a LysR-Type Transcriptional Regulator, in Listeria monocytogenes.
J. Mol. Biol., 430, 2018
|
|
5T4B
 
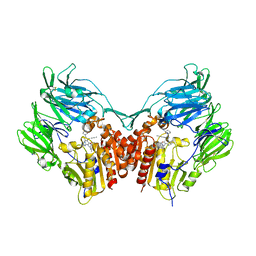 | | Human DPP4 in complex with a ligand 34a | | Descriptor: | 2-[(3R)-3-aminopiperidin-1-yl]-3-(but-2-yn-1-yl)-5-[(4-methylquinazolin-2-yl)methyl]-3H-imidazo[2,1-b]purin-4(5H)-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Scaffold-hopping from xanthines to tricyclic guanines: A case study of dipeptidyl peptidase 4 (DPP4) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
2OQ6
 
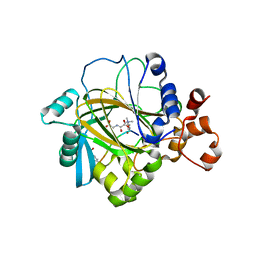 | | Crystal structure of JMJD2A complexed with histone H3 peptide trimethylated at Lys9 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Pilka, E.S, Ng, S.S, Kavanagh, K.L, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
4WZ8
 
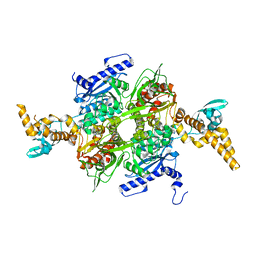 | |
6B7B
 
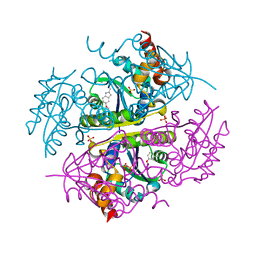 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 5-methoxy-2-methyl-1H-indole | | Descriptor: | 5-methoxy-2-methyl-1H-indole, DIMETHYL SULFOXIDE, PYROPHOSPHATE 2-, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
4X5X
 
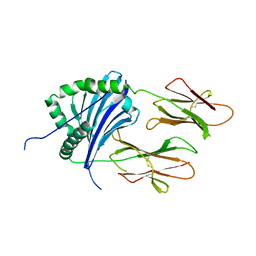 | |
6B7J
 
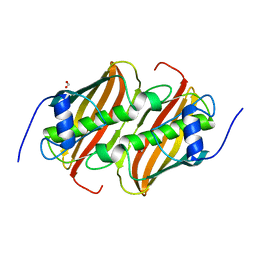 | |
6B8G
 
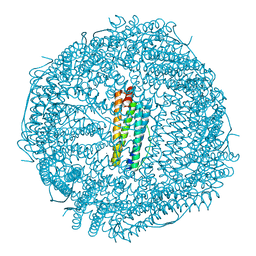 | | Twice-Contracted Human Heavy-Chain Ferritin Crystal-Hydrogel Hybrid | | Descriptor: | CALCIUM ION, FE (III) ION, Ferritin heavy chain | | Authors: | Zhang, L, Bailey, J.B, Subramanian, R, Tezcan, F.A. | | Deposit date: | 2017-10-07 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Hyperexpandable, self-healing macromolecular crystals with integrated polymer networks.
Nature, 557, 2018
|
|
2OQ7
 
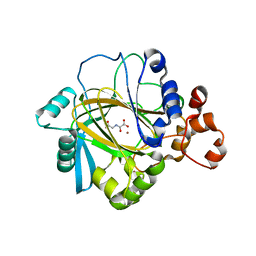 | | The crystal structure of JMJD2A complexed with Ni and N-oxalylglycine | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Kavanagh, K.L, Ng, S.S, Pilka, E, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
4WZM
 
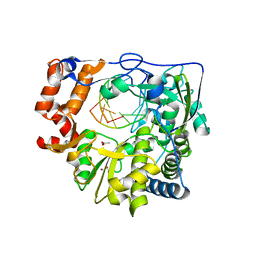 | | Mutant K18E of RNA dependent RNA polymerase from Foot-and-Mouth Disease Virus complexed with RNA | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Verdaguer, N, Ferrer-Orta, C, de la Higuera, N. | | Deposit date: | 2014-11-20 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Multifunctionality of a picornavirus polymerase domain: nuclear localization signal and nucleotide recognition.
J.Virol., 89, 2015
|
|
4WZS
 
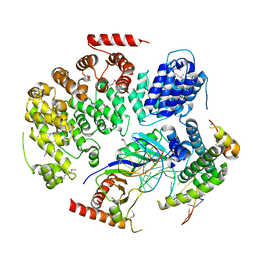 | |
6B9Y
 
 | | Trastuzumab Fab v3 in complex with 5-phenyl meditope variant | | Descriptor: | Immunoglobulin G binding protein A, Protein L, Trastuzumab Fab heavy chain, ... | | Authors: | Bzymek, K.P, King, J.D, Williams, J.C. | | Deposit date: | 2017-10-11 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Template-Catalyzed, Disulfide Conjugation of Monoclonal Antibodies Using a Natural Amino Acid Tag.
Bioconjug. Chem., 29, 2018
|
|
4X06
 
 | |
2OQV
 
 | | Human Dipeptidyl Peptidase IV (DPP4) with piperidine-constrained phenethylamine | | Descriptor: | (3R,4R)-1-{6-[3-(METHYLSULFONYL)PHENYL]PYRIMIDIN-4-YL}-4-(2,4,5-TRIFLUOROPHENYL)PIPERIDIN-3-AMINE, Dipeptidyl peptidase 4 (Dipeptidyl peptidase IV) (DPP IV) | | Authors: | Pei, Z, Li, X, von Geldern, T.W, Longenecker, K.L, Pireh, D, Stewart, K.D. | | Deposit date: | 2007-02-01 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Structure-Activity Relationships of Piperidinone- and Piperidine-Constrained Phenethylamines as Novel, Potent, and Selective Dipeptidyl Peptidase IV Inhibitors.
J.Med.Chem., 50, 2007
|
|
5TWS
 
 | | Post-catalytic complex of human Polymerase Mu (H329A) with newly incorporated UTP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2016-11-14 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
2OSU
 
 | | Probable glutaminase from Bacillus subtilis complexed with 6-diazo-5-oxo-L-norleucine | | Descriptor: | 6-DIAZENYL-5-OXO-L-NORLEUCINE, Glutaminase 1 | | Authors: | Kim, Y, Dementieva, I, Vinokour, E, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-06 | | Release date: | 2007-03-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The structure of probable glutaminase from B. subtilis complexed with its inhibitor 6-diazo-5-oxo-L-norleucine
To be Published
|
|
4WQK
 
 | | Crystal structure of aminoglycoside nucleotidylyltransferase ANT(2")-Ia, apo form | | Descriptor: | 2''-aminoglycoside nucleotidyltransferase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cox, G, Stogios, P.J, Savchenko, A, Wright, G.D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Structural and Molecular Basis for Resistance to Aminoglycoside Antibiotics by the Adenylyltransferase ANT(2)-Ia.
Mbio, 6, 2015
|
|
4WW5
 
 | | Crystal structure of binary complex Bud32-Cgi121 in complex with AMPP | | Descriptor: | ACETATE ION, EKC/KEOPS complex subunit BUD32, EKC/KEOPS complex subunit CGI121, ... | | Authors: | Zhang, W. | | Deposit date: | 2014-11-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
2OV2
 
 | | The crystal structure of the human RAC3 in complex with the CRIB domain of human p21-activated kinase 4 (PAK4) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ugochukwu, E, Yang, X, Elkins, J.M, Burgess-Brown, N, Bunkoczi, G, Debreczeni, J.E.D, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the human RAC3 in complex with the CRIB domain of human p21-activated kinase 4 (PAK4)
To be Published
|
|
6BF7
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | Descriptor: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-07 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BFY
 
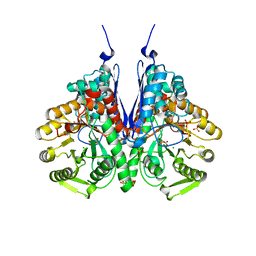 | | Crystal structure of enolase from Escherichia coli with bound 2-phosphoglycerate substrate | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, Enolase, GLYCEROL, ... | | Authors: | Erlandsen, H, Wright, D, Krucinska, J. | | Deposit date: | 2017-10-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Functional Studies of Bacterial Enolase, a Potential Target against Gram-Negative Pathogens.
Biochemistry, 58, 2019
|
|
4WW9
 
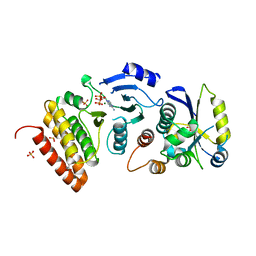 | |
6BEW
 
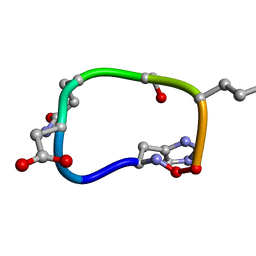 | | Solution structure of de novo macrocycle design7.2 | | Descriptor: | (DHI)P(DAS)(DGN)(DSN)(DGL)P | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | Deposit date: | 2017-10-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
4WWM
 
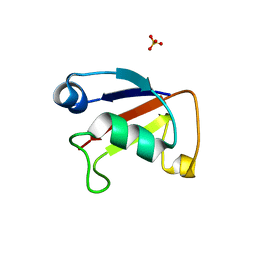 | | X-ray crystal structure of Sulfolobus solfataricus Urm1 | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Bray, S.M, Anjum, S.R, Blackwood, J.K, Kilkenny, M.L, Coelho, M.A, Foster, B.M, Pellegrini, L, Robinson, N.P. | | Deposit date: | 2014-11-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Involvement of a eukaryotic-like ubiquitin-related modifier in the proteasome pathway of the archaeon Sulfolobus acidocaldarius.
Nat Commun, 6, 2015
|
|
