1CHN
 
 | |
1CI0
 
 | | PNP OXIDASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | FLAVIN MONONUCLEOTIDE, PROTEIN (PNP OXIDASE) | | Authors: | Shi, W, Ostrov, D.A, Gerchman, S.E, Graziano, V, Kycia, H, Studier, B, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-04-06 | | Release date: | 1999-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of PNP Oxidase from S. Cerevisiae
To be Published
|
|
1D07
 
 | | Hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis UT26 with 1,3-propanediol, a product of debromidation of dibrompropane, at 2.0A resolution | | Descriptor: | 1,3-PROPANDIOL, BROMIDE ION, HALOALKANE DEHALOGENASE | | Authors: | Marek, J, Vevodova, J, Damborsky, J, Smatanova, I, Svensson, L.A, Newman, J, Nagata, Y, Takagi, M. | | Deposit date: | 1999-09-09 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26.
Biochemistry, 39, 2000
|
|
1CIJ
 
 | | HALOALKANE DEHALOGENASE SOAKED WITH HIGH CONCENTRATION OF BROMIDE | | Descriptor: | BROMIDE ION, PROTEIN (HALOALKANE DEHALOGENASE) | | Authors: | Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-03-31 | | Release date: | 1999-09-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic and kinetic evidence of a collision complex formed during halide import in haloalkane dehalogenase.
Biochemistry, 38, 1999
|
|
7WSW
 
 | | Cryo-EM structure of the Potassium channel AKT1 from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
7WMV
 
 | | Structure of human SGLT1-MAP17 complex bound with LX2761 | | Descriptor: | N-[2-(dimethylamino)ethyl]-2-methyl-2-[4-[4-[[2-methyl-5-[(2S,3R,4R,5S,6R)-6-methylsulfanyl-3,4,5-tris(oxidanyl)oxan-2-yl]phenyl]methyl]phenyl]butanoylamino]propanamide, PDZK1-interacting protein 1, Sodium/glucose cotransporter 1 | | Authors: | Chen, L, Niu, Y, Cui, W. | | Deposit date: | 2022-01-17 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism of SGLT1 inhibitors.
Nat Commun, 13, 2022
|
|
1D2C
 
 | | METHYLTRANSFERASE | | Descriptor: | PROTEIN (GLYCINE N-METHYLTRANSFERASE) | | Authors: | Huang, Y, Takusagawa, F. | | Deposit date: | 1999-09-23 | | Release date: | 1999-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
1CNC
 
 | |
1D5V
 
 | | SOLUTION STRUCTURE OF THE FORKHEAD DOMAIN OF THE ADIPOCYTE-TRANSCRIPTION FACTOR FREAC-11 (S12) | | Descriptor: | S12 TRANSCRIPTION FACTOR (FKH-14) | | Authors: | van Dongen, M.J.P, Cederberg, A, Carlsson, P, Enerback, S, Wikstrom, M. | | Deposit date: | 1999-10-12 | | Release date: | 2000-10-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the DNA-binding domain of the adipocyte-transcription factor FREAC-11.
J.Mol.Biol., 296, 2000
|
|
1CR8
 
 | |
7VYI
 
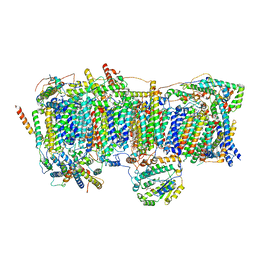 | | Membrane arm of deactive state CI from Rotenone dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-14 | | Release date: | 2022-11-23 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1D1V
 
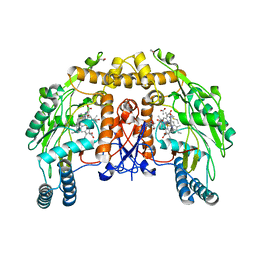 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH S-ETHYL-N-PHENYL-ISOTHIOUREA (H4B BOUND) | | Descriptor: | 2-ETHYL-1-PHENYL-ISOTHIOUREA, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-09-21 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Implications for isoform-selective inhibitor design derived from the binding mode of bulky isothioureas to the heme domain of endothelial nitric-oxide synthase.
J.Biol.Chem., 276, 2001
|
|
7VYG
 
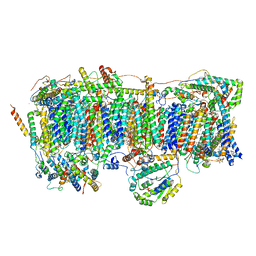 | | Membrane arm of active state CI from rotenone dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-14 | | Release date: | 2022-11-23 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1DM8
 
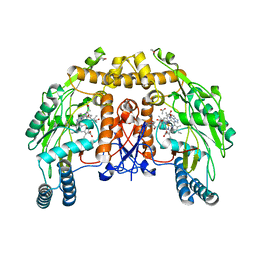 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 1,2,4-TRIAZOLE-CARBOXAMIDINE (H4B BOUND) | | Descriptor: | 1,2,4-TRIAZOLE-CARBOXAMIDINE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-12-13 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures of Bovine Endothelial Nitric Oxide Synthase Heme Domain Complexed With Various Inhibitors
To be Published
|
|
7W8F
 
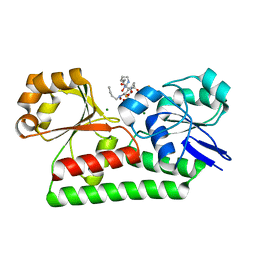 | |
7VYE
 
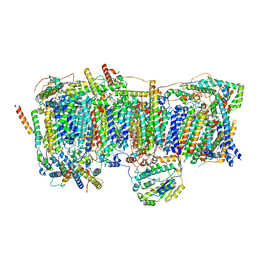 | | Membrane arm of deactive state CI from Q10-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-14 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W0H
 
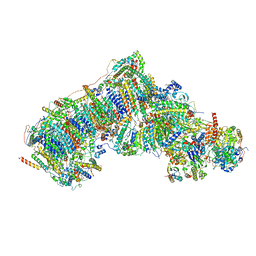 | | Deactive state CI from Q10 dataset, Subclass 2 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-12-14 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VZW
 
 | | Active state CI from Q10 dataset, Subclass 2 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-16 | | Release date: | 2022-12-14 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1DIS
 
 | | DIHYDROFOLATE REDUCTASE (E.C.1.5.1.3) COMPLEX WITH BRODIMOPRIM-4,6-DICARBOXYLATE | | Descriptor: | BRODIMOPRIM-4,6-DICARBOXYLATE, DIHYDROFOLATE REDUCTASE | | Authors: | Morgan, W.D, Birdsall, B, Polshakov, V.I, Sali, D, Kompis, I, Feeney, J. | | Deposit date: | 1995-08-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a brodimoprim analogue in its complex with Lactobacillus casei dihydrofolate reductase.
Biochemistry, 34, 1995
|
|
7VZV
 
 | | Active state CI from Q10 dataset, Subclass 1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-16 | | Release date: | 2022-12-14 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VY9
 
 | | Membrane arm of active state CI from Q10-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-13 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1DAH
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7,8-DIAMINO-NONANOIC ACID, 5'-ADENOSYL-METHYLENE-TRIPHOSPHATE, AND MANGANESE | | Descriptor: | 7,8-DIAMINO-NONANOIC ACID, DETHIOBIOTIN SYNTHETASE, MANGANESE (II) ION, ... | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
7W3E
 
 | | Bovine cytochrome c oxidese in CN-bound fully reduced state at 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A. | | Deposit date: | 2021-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic cyanide-probing for cytochrome c oxidase reveals structural bases suggesting that a putative proton transfer H-pathway pumps protons.
J.Biol.Chem., 299, 2023
|
|
7VYS
 
 | | Membrane arm of active state CI from Q1-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-15 | | Release date: | 2022-12-14 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W9Z
 
 | | Crystal structure of Bacillus subtilis YugJ in complex with NADP and nitrate | | Descriptor: | Iron-containing alcohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NITRATE ION | | Authors: | Cho, H.Y, Nam, M.S, Hong, H.J, Song, W.S, Yoon, S.I. | | Deposit date: | 2021-12-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of the Furan Aldehyde Reductase YugJ from Bacillus subtilis.
Int J Mol Sci, 23, 2022
|
|
