5OBN
 
 | | NMR solution structure of U11/U12 65K protein's C-terminal RRM domain (381-516) | | Descriptor: | RNA-binding protein 40 | | Authors: | Norppa, A.J, Kauppala, T.M, Heikkinen, H.A, Verma, B, Iwai, H, Frilander, M.J. | | Deposit date: | 2017-06-28 | | Release date: | 2018-01-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Mutations in the U11/U12-65K protein associated with isolated growth hormone deficiency lead to structural destabilization and impaired binding of U12 snRNA.
RNA, 24, 2018
|
|
1BOU
 
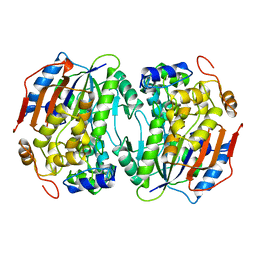 | | THREE-DIMENSIONAL STRUCTURE OF LIGAB | | Descriptor: | 4,5-DIOXYGENASE ALPHA CHAIN, 4,5-DIOXYGENASE BETA CHAIN, FE (III) ION | | Authors: | Sugimoto, K, Senda, T, Fukuda, M, Mitsui, Y. | | Deposit date: | 1998-08-06 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an aromatic ring opening dioxygenase LigAB, a protocatechuate 4,5-dioxygenase, under aerobic conditions.
Structure, 7, 1999
|
|
1BXB
 
 | | XYLOSE ISOMERASE FROM THERMUS THERMOPHILUS | | Descriptor: | XYLOSE ISOMERASE | | Authors: | Chang, C, Park, B.C, Lee, D.-S, Suh, S.W. | | Deposit date: | 1998-10-02 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of thermostable xylose isomerases from Thermus caldophilus and Thermus thermophilus: possible structural determinants of thermostability.
J.Mol.Biol., 288, 1999
|
|
6CAM
 
 | |
1GXC
 
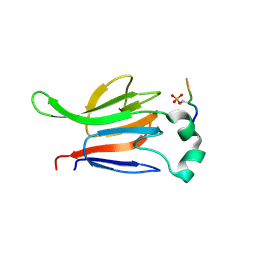 | | FHA domain from human Chk2 kinase in complex with a synthetic phosphopeptide | | Descriptor: | SERINE/THREONINE-PROTEIN KINASE CHK2, SYNTHETIC PHOSPHOPEPTIDE | | Authors: | Li, J, Williams, B.L, Haire, L.F, Goldberg, M, Wilker, E, Durocher, D, Yaffe, M.B, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-13 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Versatility of the Fha Domain in DNA-Damage Signaling by the Tumor Suppressor Kinase Chk2
Mol.Cell, 9, 2002
|
|
1H98
 
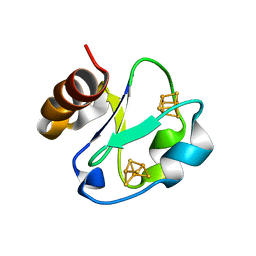 | | New Insights into Thermostability of Bacterial Ferredoxins: High Resolution Crystal Structure of the Seven-Iron Ferredoxin from Thermus thermophilus | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Macedo-Ribeiro, S, Martins, B.M, Pereira, P.J.B, Buse, G, Huber, R, Soulimane, T. | | Deposit date: | 2001-03-05 | | Release date: | 2001-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | New Insights Into the Thermostability of Bacterial Ferredoxins: High-Resolution Crystal Structure of the Seven-Iron Ferredoxin from Thermus Thermophilus
J.Biol.Inorg.Chem., 6, 2001
|
|
5OXJ
 
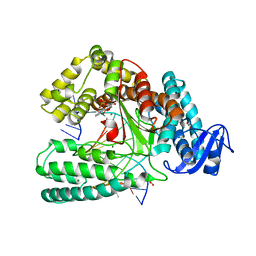 | | Crystal structure of KlenTaq mutant M747K in a closed ternary complex with a O6-MeG:BenziTP base pair | | Descriptor: | ACETATE ION, DNA polymerase I, thermostable, ... | | Authors: | Betz, K, Diederichs, K, Marx, A. | | Deposit date: | 2017-09-06 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the selective incorporation of an artificial nucleotide opposite a DNA adduct by a DNA polymerase.
Chem. Commun. (Camb.), 53, 2017
|
|
1CQ5
 
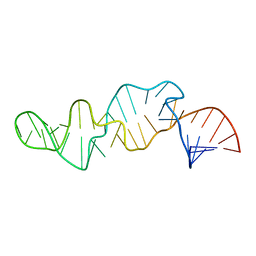 | | NMR STRUCTURE OF SRP RNA DOMAIN IV | | Descriptor: | SRP RNA DOMAIN IV | | Authors: | Schmitz, U, James, T.L, Behrens, S, Freymann, D.M, Lukavsky, P, Walter, P. | | Deposit date: | 1999-08-05 | | Release date: | 1999-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the phylogenetically most conserved domain of SRP RNA.
RNA, 5, 1999
|
|
1CXA
 
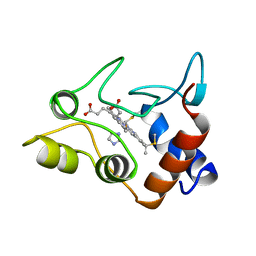 | | CRYSTALLIZATION AND X-RAY STRUCTURE DETERMINATION OF CYTOCHROME C2 FROM RHODOBACTER SPHAEROIDES IN THREE CRYSTAL FORMS | | Descriptor: | CYTOCHROME C2, HEME C, IMIDAZOLE | | Authors: | Axelrod, H.L, Feher, G, Allen, J.P, Chirino, A.J, Day, M.W, Hsu, B.T, Rees, D.C. | | Deposit date: | 1994-02-14 | | Release date: | 1995-07-10 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallization and X-ray structure determination of cytochrome c2 from Rhodobacter sphaeroides in three crystal forms.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1GV8
 
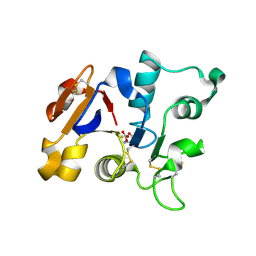 | | 18 kDa fragment of N-II domain of duck ovotransferrin | | Descriptor: | CARBONATE ION, FE (III) ION, GLYCINE, ... | | Authors: | Kuser, P, Hall, D.R, Haw, M.L, Neu, M, Lindley, P.F. | | Deposit date: | 2002-02-07 | | Release date: | 2002-02-12 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Mechanism of Iron Uptake by Transferrins: The X-Ray Structures of the 18 kDa Nii Domain Fragment of Duck Ovotransferrin and its Nitrilotriacetate Complex
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1D4L
 
 | | HIV-1 PROTEASE COMPLEXED WITH A MACROCYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | (10S,13S,1'R)-13-[1'-HYDROXY-2'-(N-P-AMINOBENZENESULFONYL-1''-AMINO-3''-METHYLBUTYL)ETHYL]-8,11-DIOXO-10-ISOPROPYL-2-OXA-9,12-DIAZABICYCLO [13.2.2]NONADECA-15,17,18-TRIENE, HIV-1 PROTEASE, SULFATE ION | | Authors: | Tyndall, J.D, Reid, R.C, Tyssen, D.P, Jardine, D.K, Todd, B, Passmore, M, March, D.R, Pattenden, L.K, Alewood, D, Hu, S.H, Alewood, P.F, Birch, C.J, Martin, J.L, Fairlie, D.P. | | Deposit date: | 1999-10-04 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Synthesis, stability, antiviral activity, and protease-bound structures of substrate-mimicking constrained macrocyclic inhibitors of HIV-1 protease.
J.Med.Chem., 43, 2000
|
|
1D5C
 
 | | CRYSTAL STRUCTURE OF PLASMODIUM FALCIPARUM RAB6 COMPLEXED WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAB6 GTPASE | | Authors: | Chattopadhyay, D, Langsley, G, Carson, M, Recacha, R, DeLucas, L, Smith, C. | | Deposit date: | 1999-10-06 | | Release date: | 2000-08-30 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the nucleotide-binding domain of Plasmodium falciparum rab6 in the GDP-bound form.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1FIO
 
 | | CRYSTAL STRUCTURE OF YEAST T-SNARE PROTEIN SSO1 | | Descriptor: | SSO1 PROTEIN, ZINC ION | | Authors: | Munson, M, Chen, X, Cocina, A.E, Schultz, S.M, Hughson, F.M. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interactions within the yeast t-SNARE Sso1p that control SNARE complex assembly.
Nat.Struct.Biol., 7, 2000
|
|
6BNR
 
 | | Carbonmonoxy hemoglobin in complex with the antisickling agent 5-methoxy-2-(pyridin-2-ylmethoxy)benzaldehyde (INN310) | | Descriptor: | 2-[(4-methoxy-2-methylphenoxy)methyl]pyridine, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Pagare, P.P, Musayev, F.N, Safo, M.K. | | Deposit date: | 2017-11-17 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational design of pyridyl derivatives of vanillin for the treatment of sickle cell disease.
Bioorg. Med. Chem., 26, 2018
|
|
6BZX
 
 | | Structure of the artificial complex alpha-Rep/Octarellin V.1 crystallized by counter diffusion in a capillary | | Descriptor: | Octarellin V.1, SODIUM ION, alpha-Rep | | Authors: | Aedo, F, Contreras-Martel, C, Martinez-Oyanedel, J, Bunster, M, Minard, P, Van de Weerdt, C, Figueroa, M. | | Deposit date: | 2017-12-26 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.107 Å) | | Cite: | Crystallization of the artificial complex alpha-Rep/Octarellin V.1 by counter diffusion allowed to have a most complete structure
To Be Published
|
|
1CFM
 
 | | CYTOCHROME F FROM CHLAMYDOMONAS REINHARDTII | | Descriptor: | CYTOCHROME F, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chi, Y.I, Huang, L.S, Zhang, Z, Fernandez-Velasco, J.G, Malkin, R, Berry, E.A. | | Deposit date: | 1998-09-18 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of a truncated form of cytochrome f from chlamydomonas reinhardtii.
Biochemistry, 39, 2000
|
|
1CHV
 
 | |
1FP2
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ISOFLAVONE O-METHYLTRANSFERASE | | Descriptor: | 4'-HYDROXY-7-METHOXYISOFLAVONE, ISOFLAVONE O-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zubieta, C, Dixon, R.A, Noel, J.P. | | Deposit date: | 2000-08-29 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases.
Nat.Struct.Biol., 8, 2001
|
|
1FMM
 
 | | SOLUTION STRUCTURE OF NFGF-1 | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Arunkumar, A.I, Srisailam, S, Kumar, T.K.S, Chiu, I.M, Yu, C. | | Deposit date: | 2000-08-18 | | Release date: | 2001-08-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and stability of an acidic fibroblast growth factor from Notophthalmus viridescens.
J.Biol.Chem., 277, 2002
|
|
1TQ1
 
 | | Solution structure of At5g66040, a putative protein from Arabidosis Thaliana | | Descriptor: | senescence-associated family protein | | Authors: | Cornilescu, C.C, Cornilescu, G, Singh, S, Lee, M.S, Tyler, E.M, Shahan, M.N, Vinarov, D, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-06-16 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a single-domain thiosulfate sulfurtransferase from Arabidopsis thaliana.
Protein Sci., 15, 2006
|
|
1FA3
 
 | |
6C3L
 
 | | Crystal structure of BCL6 BTB domain with compound 15f | | Descriptor: | B-cell lymphoma 6 protein, N-[2-(1H-indol-3-yl)ethyl]-N'-{3-[(4-methylpiperazin-1-yl)methyl]-1-[2-(morpholin-4-yl)-2-oxoethyl]-1H-indol-6-yl}thiourea | | Authors: | Linhares, B, Cheng, H, Cierpicki, T, Xue, F. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.46092153 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
1AFE
 
 | | HUMAN ALPHA-THROMBIN INHIBITION BY CBZ-PRO-AZALYS-ONP | | Descriptor: | 2-[N'-(4-AMINO-BUTYL)-HYDRAZINOCARBONYL]-PYRROLIDINE-1-CARBOXYLIC ACID BENZYL ESTER, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ... | | Authors: | De Simone, G, Balliano, G, Milla, P, Gallina, C, Giordano, C, Tarricone, C, Rizzi, M, Bolognesi, M, Ascenzi, P. | | Deposit date: | 1997-03-06 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human alpha-thrombin inhibition by the highly selective compounds N-ethoxycarbonyl-D-Phe-Pro-alpha-azaLys p-nitrophenyl ester and N-carbobenzoxy-Pro-alpha-azaLys p-nitrophenyl ester: a kinetic, thermodynamic and X-ray crystallographic study.
J.Mol.Biol., 269, 1997
|
|
1AAN
 
 | | CRYSTAL STRUCTURE ANALYSIS OF AMICYANIN AND APOAMICYANIN FROM PARACOCCUS DENITRIFICANS AT 2.0 ANGSTROMS AND 1.8 ANGSTROMS RESOLUTION | | Descriptor: | AMICYANIN, COPPER (II) ION | | Authors: | Chen, L, Durley, R.C.E, Lim, L.W, Mathews, F.S. | | Deposit date: | 1992-04-09 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis of amicyanin and apoamicyanin from Paracoccus denitrificans at 2.0 A and 1.8 A resolution.
Protein Sci., 2, 1993
|
|
5YY8
 
 | |
