1G7M
 
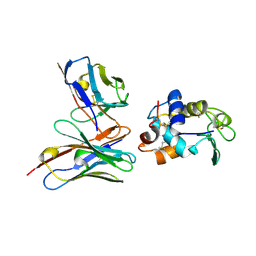 | | CRYSTAL STRUCTURE OF HEN EGG WHITE LYSOZYME (HEL) COMPLEXED WITH THE MUTANT ANTI-HEL MONOCLONAL ANTIBODY D1.3 (VLW92V) | | Descriptor: | ANTI-HEN EGG WHITE LYSOZYME MONOCLONAL ANTIBODY D1.3, LYSOZYME C | | Authors: | Sundberg, E.J, Urrutia, M, Braden, B.C, Isern, J, Mariuzza, R.A. | | Deposit date: | 2000-11-10 | | Release date: | 2000-11-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Estimation of the hydrophobic effect in an antigen-antibody protein-protein interface.
Biochemistry, 39, 2000
|
|
6BNV
 
 | |
7K7K
 
 | |
7K9L
 
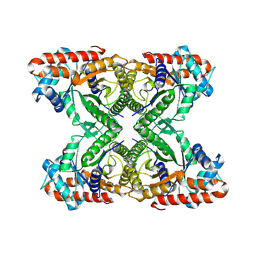 | | Aldolase, rabbit muscle (no beam-tilt refinement) | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Cianfrocco, M.A, Kearns, S.E, Cash, J.N, Li, Y. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | High-resolution cryo-EM using beam-image shift at 200 keV.
Iucrj, 7, 2020
|
|
1KZE
 
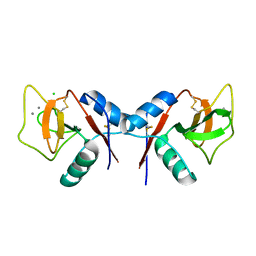 | | Complex of MBP-C and bivalent Man-terminated glycopeptide | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN C, ... | | Authors: | Ng, K.K, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
1LIE
 
 | |
6BGX
 
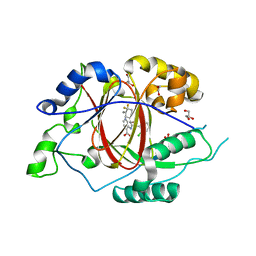 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)((4,4-difluorocyclohexyl)methoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound N42) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[(4,4-difluorocyclohexyl)methoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH8
 
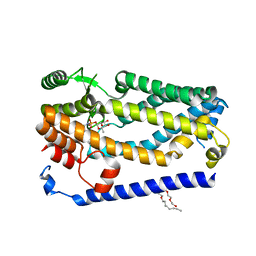 | | Crystal structure of ZMPSTE24 in complex with phosphoramidon | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CAAX prenyl protease 1 homolog, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | Goblirsch, B.R, Arachea, B.T, Wiener, M.C. | | Deposit date: | 2017-10-30 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Phosphoramidon inhibits the integral membrane protein zinc metalloprotease ZMPSTE24.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1GA2
 
 | | THE CRYSTAL STRUCTURE OF ENDOGLUCANASE 9G FROM CLOSTRIDIUM CELLULOLYTICUM COMPLEXED WITH CELLOBIOSE | | Descriptor: | ACETIC ACID, CALCIUM ION, ENDOGLUCANASE 9G, ... | | Authors: | Mandelman, D, Belaich, A, Belaich, J.P, Aghajari, N, Driguez, H, Haser, R. | | Deposit date: | 2000-11-29 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-Ray Crystal Structure of the Multidomain Endoglucanase Cel9G from Clostridium
cellulolyticum Complexed with Natural and Synthetic Cello-Oligosaccharides
J.BACTERIOL., 185, 2003
|
|
7KDN
 
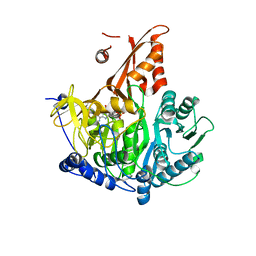 | |
1FWP
 
 | |
7JL2
 
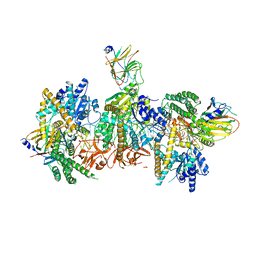 | | Cryo-EM structure of MDA5-dsRNA filament in complex with TRIM65 PSpry domain (Trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JRG
 
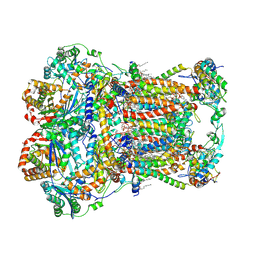 | | Plant Mitochondrial complex III2 from Vigna radiata | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Alpha-MPP, ... | | Authors: | Maldonado, M, Letts, J.A. | | Deposit date: | 2020-08-12 | | Release date: | 2021-01-20 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Atomic structures of respiratory complex III 2 , complex IV, and supercomplex III 2 -IV from vascular plants.
Elife, 10, 2021
|
|
1L7R
 
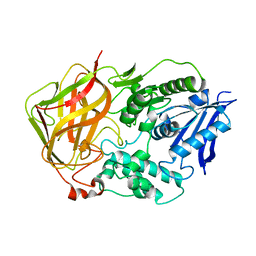 | | Tyr44Phe Mutant of Bacterial Cocaine Esterase cocE | | Descriptor: | cocaine esterase | | Authors: | Turner, J.M, Larsen, N.A, Basran, A, Barbas III, C.F, Bruce, N.C, Wilson, I.A, Lerner, R.A. | | Deposit date: | 2002-03-16 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Biochemical characterization and structural analysis of a highly proficient cocaine
esterase.
Biochemistry, 41, 2002
|
|
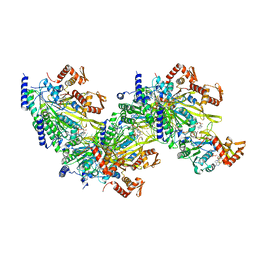
7JY8
 
 | |
1LA4
 
 | | Solution Structure of SGTx1 | | Descriptor: | SGTx1 | | Authors: | Lee, C.W, Roh, S.H, Kim, S, Endoh, H, Kodera, Y, Maeda, T, Swartz, K.J, Kim, J.I. | | Deposit date: | 2002-03-28 | | Release date: | 2003-11-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Functional Characterization of SGTx1, a Modifier of Kv2.1 Channel Gating
Biochemistry, 43, 2004
|
|
7JSF
 
 | | Adeno-Associated Virus Helicase domain Heptamer with ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*T)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
1LB3
 
 | | Structure of recombinant mouse L chain ferritin at 1.2 A resolution | | Descriptor: | CADMIUM ION, FERRITIN LIGHT CHAIN 1, GLYCEROL, ... | | Authors: | Granier, T, Langlois D'Estaintot, B, Gallois, B, Chevalier, J.-M, Precigoux, G, Santambrogio, P, Arosio, P. | | Deposit date: | 2002-04-02 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural description of the active sites of mouse L-chain ferritin at 1.2A resolution
J.Biol.Inorg.Chem., 8, 2003
|
|
6BYT
 
 | | Complex structure of LOR107 mutant (R320) with tetrasaccharide substrate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Ulaganathan, T.S, Cygler, M. | | Deposit date: | 2017-12-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-function analyses of a PL24 family ulvan lyase reveal key features and suggest its catalytic mechanism.
J. Biol. Chem., 293, 2018
|
|
6BGY
 
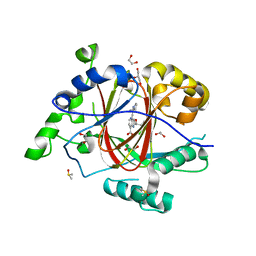 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(1-methylpyrrolidin-2-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound 46) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(S)-(2-chlorophenyl){2-[(2R)-1-methylpyrrolidin-2-yl]ethoxy}methyl]-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
1LL6
 
 | |
1HXR
 
 | | CRYSTAL STRUCTURE OF MSS4 AT 1.65 ANGSTROMS | | Descriptor: | GUANINE NUCLEOTIDE EXCHANGE FACTOR MSS4, ZINC ION | | Authors: | Zhu, Z, Dumas, J.J, Lietzke, S.E, Lambright, D.G. | | Deposit date: | 2001-01-16 | | Release date: | 2001-02-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A helical turn motif in Mss4 is a critical determinant of Rab binding and nucleotide release.
Biochemistry, 40, 2001
|
|
6BU3
 
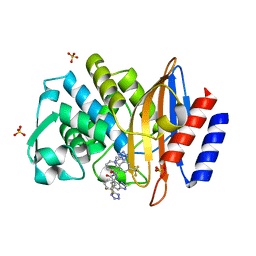 | |
7JPE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with m7GpppA Cap-0 and SAM Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Non-structural protein 10, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-07 | | Release date: | 2020-08-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1LLO
 
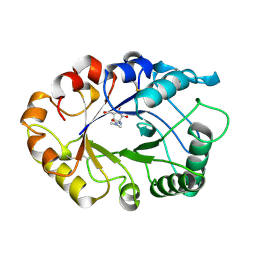 | | HEVAMINE A (A PLANT ENDOCHITINASE/LYSOZYME) COMPLEXED WITH ALLOSAMIDIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, Hevamine-A | | Authors: | Terwisscha Van Scheltinga, A.C, Armand, S, Kalk, K.H, Isogai, A, Henrissat, B, Dijkstra, B.W. | | Deposit date: | 1995-11-08 | | Release date: | 1996-03-08 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Stereochemistry of chitin hydrolysis by a plant chitinase/lysozyme and X-ray structure of a complex with allosamidin: evidence for substrate assisted catalysis.
Biochemistry, 34, 1995
|
|
