3KRL
 
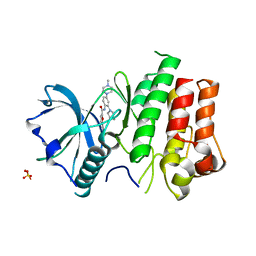 | | cFMS Tyrosine kinase in complex with 5-Cyano-furan-2-carboxylic acid [4-(4-methyl-piperazin-1-yl)-2-piperidin-1-yl-phenyl]-amide | | Descriptor: | 5-cyano-N-[4-(4-methylpiperazin-1-yl)-2-piperidin-1-ylphenyl]furan-2-carboxamide, Macrophage colony-stimulating factor 1 receptor, Basic fibroblast growth factor receptor 1, ... | | Authors: | Schubert, C. | | Deposit date: | 2009-11-18 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of a Potent Class of Arylamide Colony-Stimulating Factor-1 Receptor Inhibitors Leading to Anti-inflammatory Clinical Candidate 4-Cyano-N-[2-(1-cyclohexen-1-yl)-4-[1-[(dimethylamino)acetyl]-4-piperidinyl]phenyl]-1H-imidazole-2-carboxamide (JNJ-28312141).
J.Med.Chem., 54, 2011
|
|
4D1G
 
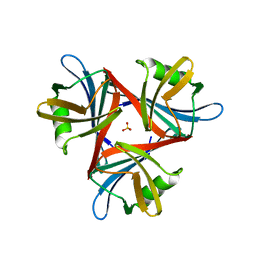 | | Crystal structure of the fiber head domain of the Atadenovirus snake adenovirus 1, native, second P212121 crystal form | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, FIBER PROTEIN, SULFATE ION | | Authors: | Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-05-01 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the fibre head domain of the Atadenovirus Snake Adenovirus 1.
PLoS ONE, 9, 2014
|
|
5VP1
 
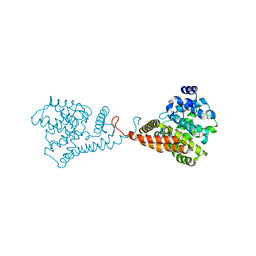 | | Discovery of Clinical Candidate N-{(1S)-1-[3-Fluoro-4-(trifluoromethoxy)phenyl]-2-methoxyethyl}-7-methoxy-2-oxo-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide (TAK-915), A Highly Potent, Selective, and Brain-Penetrating Phosphodiesterase 2A Inhibitor for the Treatment of Cognitive Disorders | | Descriptor: | MAGNESIUM ION, N-{(1S)-2-hydroxy-2-methyl-1-[4-(trifluoromethoxy)phenyl]propyl}-6-methyl-5-(3-methyl-1H-1,2,4-triazol-1-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide, ZINC ION, ... | | Authors: | Hoffman, I.D. | | Deposit date: | 2017-05-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of a Novel Series of Pyrazolo[1,5-a]pyrimidine-Based Phosphodiesterase 2A Inhibitors Structurally Different from N-((1S)-1-(3-Fluoro-4-(trifluoromethoxy)phenyl)-2-methoxyethyl)-7-methoxy-2-oxo-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide (TAK-915), for the Treatment of Cognitive Disorders.
Chem. Pharm. Bull., 65, 2017
|
|
2QA9
 
 | |
1IKW
 
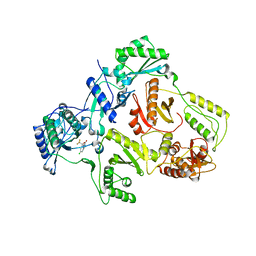 | | Wild Type HIV-1 Reverse Transcriptase in Complex with Efavirenz | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, POL POLYPROTEIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2001-05-07 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant.
Eur.J.Biochem., 269, 2002
|
|
1T7I
 
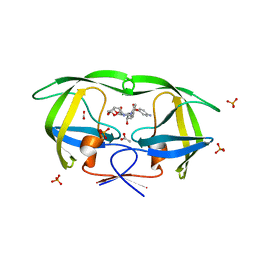 | | The structural and thermodynamic basis for the binding of TMC114, a next-generation HIV-1 protease inhibitor. | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | King, N.M, Prabu-Jeyabalan, M, Nalivaika, E.A, Wigerinck, P.B.T.P, De Bethune, M.-P, Schiffer, C.A. | | Deposit date: | 2004-05-10 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor
J.Med.Chem., 48, 2005
|
|
1YA4
 
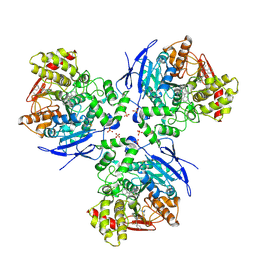 | | Crystal Structure of Human Liver Carboxylesterase 1 in complex with tamoxifen | | Descriptor: | (Z)-2-[4-(1,2)-DIPHENYL-1-BUTENYL)-PHENOXY]-N,N-DIMETHYLETHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CES1 protein, ... | | Authors: | Fleming, C.D, Bencharit, S, Edwards, C.C, Hyatt, J.L, Morton, C.L, Howard-Williams, E.L, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2004-12-17 | | Release date: | 2005-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into drug processing by human carboxylesterase 1: tamoxifen, mevastatin, and inhibition by benzil.
J.Mol.Biol., 352, 2005
|
|
3O9G
 
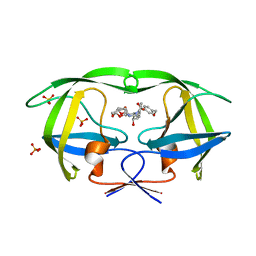 | | Crystal Structure of wild-type HIV-1 Protease in complex with af53 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-1-benzyl-3-[(2-ethylbutyl){[4-(hydroxymethyl)phenyl]sulfonyl}amino]-2-hydroxypropyl}carbamate, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
1HVO
 
 | |
1HTG
 
 | |
1M4W
 
 | | Thermophilic b-1,4-xylanase from Nonomuraea flexuosa | | Descriptor: | ACETATE ION, GLYCEROL, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hakulinen, N, Turunen, O, Janis, J, Leisola, M, Rouvinen, J. | | Deposit date: | 2002-07-05 | | Release date: | 2003-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structures of thermophilic beta-1,4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa. Comparison of twelve xylanases in relation to their thermal stability.
Eur.J.Biochem., 270, 2003
|
|
1UUI
 
 | | NMR structure of a synthetic small molecule, rbt158, bound to HIV-1 TAR RNA | | Descriptor: | 4-[AMINO(IMINO)METHYL]-1-[2-(3-AMMONIOPROPOXY)-5-METHOXYBENZYL]PIPERAZIN-1-IUM, 5'-R(*GP*GP*CP*AP*GP*AP*UP*CP*UP*GP*AP*GP*CP* CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*UP*CP*UP*GP*CP*C)-3' | | Authors: | Davis, B, Afshar, M, Varani, G, Karn, J, Murchie, A.I.H, Lentzen, G, Drysdale, M.J, Potter, A.J, Bower, J, Aboul-Ela, F. | | Deposit date: | 2003-12-19 | | Release date: | 2004-02-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational Design of Inhibitors of HIV-1 Tar RNA Through the Stabilisation of Electrostatic "Hot Spots"
J.Mol.Biol., 336, 2004
|
|
4HWW
 
 | | Crystal structure of human Arginase-1 complexed with inhibitor 9 | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5R)-5-amino-5-carboxy-7-(piperidin-1-yl)heptyl](trihydroxy)borate(1-) | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D.L, Golebiowski, A, Ji, M, Zhang, M, Beckett, P, Sheeler, R, Andreoli, M, Conway, B, Mahboubi, K, Schroeter, H, Van Zandt, M.C, Podjarny, A. | | Deposit date: | 2012-11-09 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Discovery of (R)-2-Amino-6-borono-2-(2-(piperidin-1-yl)ethyl)hexanoic Acid and Congeners As Highly Potent Inhibitors of Human Arginases I and II for Treatment of Myocardial Reperfusion Injury.
J.Med.Chem., 56, 2013
|
|
1WBM
 
 | | HIV-1 protease in complex with symmetric inhibitor, BEA450 | | Descriptor: | (2R,3R,4R,5R)-3,4-DIHYDROXY-N,N'-BIS[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]-2,5-BIS(2-PHENYLETHYL)HEXANEDIAMIDE, POL PROTEIN (FRAGMENT) | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2004-11-02 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease in Complex with Symmetric Inhibitor, Bea450
To be Published
|
|
4HYS
 
 | |
4HYU
 
 | |
3HR9
 
 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase F40I mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-glucanase, ... | | Authors: | Tsai, L.C, Huang, H.C, Hsiao, C.H. | | Deposit date: | 2009-06-09 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase mutant F40I
To be Published
|
|
4I53
 
 | |
4INB
 
 | | Crystal Structure of the N-Terminal Domain of HIV-1 Capsid in Complex With benzodiazepine Inhibitor | | Descriptor: | (3Z)-3-{[(2-methoxyethyl)amino]methylidene}-1-methyl-5-phenyl-7-(trifluoromethyl)-1H-1,5-benzodiazepine-2,4(3H,5H)-dione, Gag protein, SODIUM ION | | Authors: | Coulombe, R. | | Deposit date: | 2013-01-04 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Monitoring Binding of HIV-1 Capsid Assembly Inhibitors Using (19) F Ligand-and (15) N Protein-Based NMR and X-ray Crystallography: Early Hit Validation of a Benzodiazepine Series.
Chemmedchem, 8, 2013
|
|
2AZB
 
 | | HIV-1 Protease NL4-3 3X mutant in complex with inhibitor, TL-3 | | Descriptor: | PROTEASE RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Kutilek, V, Morris, G.M, Lin, Y.-C, Elder, J.H, Torbett, B.E, Stout, C.D. | | Deposit date: | 2005-09-09 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights into the Mechanisms of Drug Resistance in HIV-1 Protease NL4-3
J.Mol.Biol., 356, 2006
|
|
3PS4
 
 | | PDZ domain from Human microtubule-associated serine/threonine-protein kinase 1 | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Microtubule-associated serine/threonine-protein kinase 1 | | Authors: | Ugochukwu, E, Wang, J, Krojer, T, Muniz, J.R.C, Sethi, R, Pike, A.C.W, Roos, A, Salah, E, Cocking, R, Savitsky, P, Doyle, D.A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-30 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PDZ domain from Human microtubule-associated serine/threonine-protein kinase 1
TO BE PUBLISHED
|
|
3Q94
 
 | | The crystal structure of fructose 1,6-bisphosphate aldolase from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, ACETATE ION, Fructose-bisphosphate aldolase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-07 | | Release date: | 2011-01-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | The crystal structure of fructose 1,6-bisphosphate aldolase from Bacillus anthracis str. 'Ames Ancestor'.
To be Published
|
|
2AZ9
 
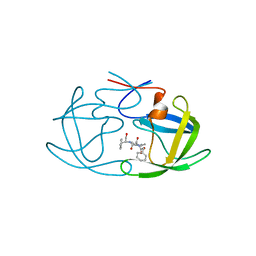 | | HIV-1 Protease NL4-3 1X mutant | | Descriptor: | PROTEASE RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Kutilek, V, Morris, G.M, Lin, Y.-C, Elder, J.H, Torbett, B.E, Stout, C.D. | | Deposit date: | 2005-09-09 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Mechanisms of Drug Resistance in HIV-1 Protease NL4-3
J.Mol.Biol., 356, 2006
|
|
2AOG
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a substrate analog P2-NC | | Descriptor: | ACETIC ACID, GLYCEROL, HIV-1 PROTEASE (RETROPEPSIN), ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOD
 
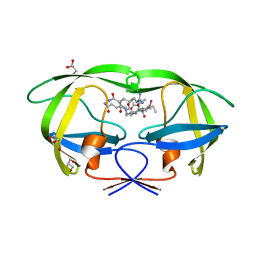 | | Crystal structure analysis of HIV-1 protease with a substrate analog P2-NC | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, HIV-1 PROTEASE, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
