2CDU
 
 | | The Crystal Structure of Water-forming NAD(P)H Oxidase from Lactobacillus sanfranciscensis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH OXIDASE | | Authors: | Lountos, G.T, Jiang, R, Wellborn, W.B, Thaler, T.L, Bommarius, A.S, Orville, A.M. | | Deposit date: | 2006-01-28 | | Release date: | 2006-07-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Nad(P)H Oxidase from Lactobacillus Sanfranciscensis: Insights Into the Conversion of O(2) Into Two Water Molecules by the Flavoenzyme.
Biochemistry, 45, 2006
|
|
1XY2
 
 | | CRYSTAL STRUCTURE ANALYSIS OF DEAMINO-OXYTOCIN. CONFORMATIONAL FLEXIBILITY AND RECEPTOR BINDING | | Descriptor: | OXYTOCIN | | Authors: | Cooper, S, Blundell, T.L, Pitts, J.E, Wood, S.P, Tickle, I.J. | | Deposit date: | 1987-06-05 | | Release date: | 1988-04-16 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure analysis of deamino-oxytocin: conformational flexibility and receptor binding.
Science, 232, 1986
|
|
1FK9
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Milton, J, Weaver, K.L, Short, S.A, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-08-09 | | Release date: | 2000-11-03 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase.
STRUCTURE FOLD.DES., 8, 2000
|
|
197L
 
 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | Deposit date: | 1995-11-06 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
1FKP
 
 | | CRYSTAL STRUCTURE OF NNRTI RESISTANT K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Milton, J, Weaver, K.L, Short, S.A, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-08-10 | | Release date: | 2000-11-03 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase.
Structure Fold.Des., 8, 2000
|
|
2CJJ
 
 | | Crystal Structure of the MYB domain of the RAD transcription factor from Antirrhinum majus | | Descriptor: | RADIALIS | | Authors: | Stevenson, C.E.M, Burton, N, Costa, M.M, Nath, U, Dixon, R.A, Coen, E.S, Lawson, D.M. | | Deposit date: | 2006-04-04 | | Release date: | 2006-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Myb Domain of the Rad Transcription Factor from Antirrhinum Majus.
Proteins: Struct., Funct., Bioinf., 65, 2006
|
|
1FS4
 
 | | Structures of glycogen phosphorylase-inhibitor complexes and the implications for structure-based drug design | | Descriptor: | 1-DEOXY-1-METHOXYCARBAMIDO-BETA-D-GLUCO-2-HEPTULOPYRANOSONAMIDE, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-08 | | Release date: | 2000-10-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
1FAK
 
 | | HUMAN TISSUE FACTOR COMPLEXED WITH COAGULATION FACTOR VIIA INHIBITED WITH A BPTI-MUTANT | | Descriptor: | CALCIUM ION, PROTEIN (5L15), PROTEIN (BLOOD COAGULATION FACTOR VIIA), ... | | Authors: | Zhang, E, St Charles, R, Tulinsky, A. | | Deposit date: | 1998-12-28 | | Release date: | 1999-12-03 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of extracellular tissue factor complexed with factor VIIa inhibited with a BPTI mutant.
J.Mol.Biol., 285, 1999
|
|
2CPO
 
 | | CHLOROPEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLOROPEROXIDASE, ... | | Authors: | Sundaramoorthy, M, Poulos, T.L. | | Deposit date: | 1996-02-10 | | Release date: | 1997-02-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of chloroperoxidase: a heme peroxidase--cytochrome P450 functional hybrid.
Structure, 3, 1995
|
|
1FOQ
 
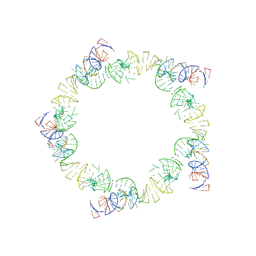 | | PENTAMERIC MODEL OF THE BACTERIOPHAGE PHI29 PROHEAD RNA | | Descriptor: | BACTERIOPHAGE PHI29 PROHEAD RNA | | Authors: | Simpson, A.A, Tao, Y, Leiman, P.G, Badasso, M.O, He, Y, Jardine, P.J, Olson, N.H, Morais, M.C, Grimes, S, Anderson, D.L, Baker, T.S, Rossmann, M.G. | | Deposit date: | 2000-08-28 | | Release date: | 2000-12-22 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structure of the bacteriophage phi29 DNA packaging motor.
Nature, 408, 2000
|
|
1FT4
 
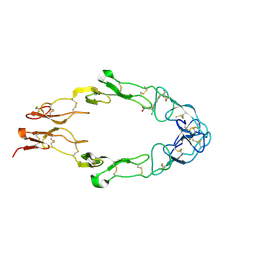 | |
1HL8
 
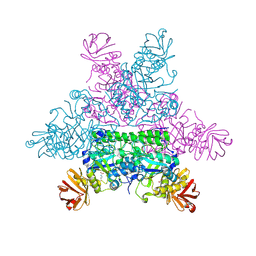 | |
2OQE
 
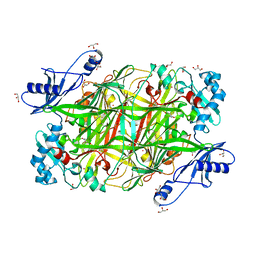 | |
1YI8
 
 | |
3R45
 
 | | Structure of a CENP-A-Histone H4 Heterodimer in complex with chaperone HJURP | | Descriptor: | GLYCEROL, Histone H3-like centromeric protein A, Histone H4, ... | | Authors: | Hu, H, Liu, Y, Wang, M, Fang, J, Huang, H, Yang, N, Li, Y, Wang, J, Yao, X, Shi, Y, Li, G, Xu, R.M. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP
Genes Dev., 25, 2011
|
|
1HYR
 
 | |
1Z38
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with inosine | | Descriptor: | INOSINE, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
2OOV
 
 | |
2F8Q
 
 | | An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 | | Descriptor: | MAGNESIUM ION, alkaline thermostable endoxylanase | | Authors: | Ramakumar, S, Manikandan, K, Bhardwaj, A, Ghosh, A, Reddy, V.S. | | Deposit date: | 2005-12-03 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of native and xylosaccharide-bound alkali thermostable xylanase from an alkalophilic Bacillus sp. NG-27: structural insights into alkalophilicity and implications for adaptation to polyextreme conditions.
Protein Sci., 15, 2006
|
|
2HLP
 
 | | CRYSTAL STRUCTURE OF THE E267R MUTANT OF A HALOPHILIC MALATE DEHYDROGENASE IN THE APO FORM | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, SODIUM ION | | Authors: | Richard, S.B, Madern, D, Garcin, E, Zaccai, G. | | Deposit date: | 1999-04-23 | | Release date: | 2000-02-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Halophilic adaptation: novel solvent protein interactions observed in the 2.9 and 2.6 A resolution structures of the wild type and a mutant of malate dehydrogenase from Haloarcula marismortui.
Biochemistry, 39, 2000
|
|
1AD1
 
 | | DIHYDROPTEROATE SYNTHETASE (APO FORM) FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIHYDROPTEROATE SYNTHETASE, POTASSIUM ION | | Authors: | Kostrewa, D, Oefner, C, D'Arcy, A. | | Deposit date: | 1997-02-19 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of the dihydropteroate synthase from Staphylococcus aureus.
J.Mol.Biol., 268, 1997
|
|
1AD4
 
 | |
1Z2W
 
 | | Crystal structure of mouse Vps29 complexed with Mn2+ | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Vacuolar protein sorting 29 | | Authors: | Collins, B.M, Skinner, C.F, Watson, P.J, Seaman, M.N.J, Owen, D.J. | | Deposit date: | 2005-03-10 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly
NAT.STRUCT.MOL.BIOL., 12, 2005
|
|
1Z35
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2-fluoroadenosine | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1AA3
 
 | | C-TERMINAL DOMAIN OF THE E. COLI RECA, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RECA | | Authors: | Aihara, H, Ito, Y, Kurumizaka, H, Terada, T, Yokoyama, S, Shibata, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-01-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | An interaction between a specified surface of the C-terminal domain of RecA protein and double-stranded DNA for homologous pairing.
J.Mol.Biol., 274, 1997
|
|
