2DJG
 
 | | Re-determination of the native structure of human dipeptidyl peptidase I (cathepsin C) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Dipeptidyl-peptidase 1, ... | | Authors: | Molgaard, A, Arnau, J, Lauritzen, C, Larsen, S, Petersen, G, Pedersen, J. | | Deposit date: | 2006-04-02 | | Release date: | 2006-11-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structure of human dipeptidyl peptidase I (cathepsin C) in complex with the inhibitor Gly-Phe-CHN2
Biochem.J., 401, 2007
|
|
1TOP
 
 | |
1CRC
 
 | | CYTOCHROME C AT LOW IONIC STRENGTH | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Sanishvili, R, Volz, K.W, Westbrook, E.M, Margoliash, E. | | Deposit date: | 1995-03-22 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The low ionic strength crystal structure of horse cytochrome c at 2.1 A resolution and comparison with its high ionic strength counterpart.
Structure, 3, 1995
|
|
2OT4
 
 | | Structure of a hexameric multiheme c nitrite reductase from the extremophile bacterium Thiolkalivibrio nitratireducens | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2007-02-07 | | Release date: | 2008-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens.
J.Mol.Biol., 389, 2009
|
|
4QDQ
 
 | | Physical basis for Nrp2 ligand binding | | Descriptor: | GLYCEROL, Neuropilin-2, SULFATE ION | | Authors: | Parker, M.W, Vander Kooi, C.W. | | Deposit date: | 2014-05-14 | | Release date: | 2015-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for VEGF-C Binding to Neuropilin-2 and Sequestration by a Soluble Splice Form.
Structure, 23, 2015
|
|
3ZIW
 
 | | Clostridium perfringens enterotoxin, D48A mutation and N-terminal 37 residues deleted | | Descriptor: | HEAT-LABILE ENTEROTOXIN B CHAIN, HEXAETHYLENE GLYCOL | | Authors: | Yelland, T, Naylor, C.E, Savva, C.G, Basak, A.K. | | Deposit date: | 2013-01-14 | | Release date: | 2014-01-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a C. Perfringens Enterotoxin Mutant in Complex with a Modified Claudin-2 Extracellular Loop 2
J.Mol.Biol., 426, 2014
|
|
2ZYS
 
 | | A. Fulgidus lipase with fatty acid fragment and chloride | | Descriptor: | CHLORIDE ION, Lipase, putative, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
2ZYI
 
 | | A. Fulgidus lipase with fatty acid fragment and calcium | | Descriptor: | CALCIUM ION, Lipase, putative, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-22 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
2ZYH
 
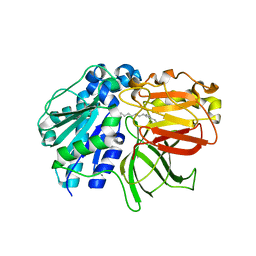 | | mutant A. Fulgidus lipase S136A complexed with fatty acid fragment | | Descriptor: | CALCIUM ION, HEXADECANE, Lipase, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-22 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
1HRC
 
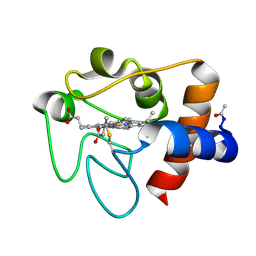 | |
4R75
 
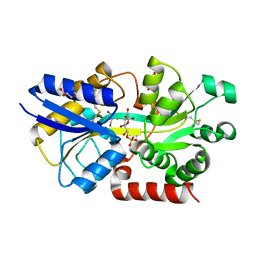 | | Structure of the periplasmic binding protein AfuA from Actinobacillus pleuropneumoniae (exogenous sedoheptulose-7-phosphate bound) | | Descriptor: | 1-C-(hydroxymethyl)-6-O-phosphono-beta-D-altrofuranose, ABC-type Fe3+ transport system, periplasmic component, ... | | Authors: | Sit, B, Calmettes, C, Moraes, T.F. | | Deposit date: | 2014-08-26 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.278 Å) | | Cite: | Active Transport of Phosphorylated Carbohydrates Promotes Intestinal Colonization and Transmission of a Bacterial Pathogen.
Plos Pathog., 11, 2015
|
|
1LC1
 
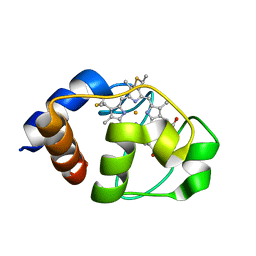 | |
1EXQ
 
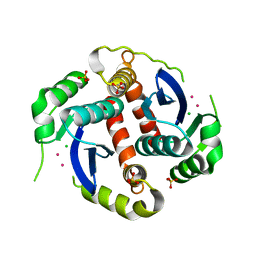 | | CRYSTAL STRUCTURE OF THE HIV-1 INTEGRASE CATALYTIC CORE DOMAIN | | Descriptor: | CADMIUM ION, CHLORIDE ION, POL POLYPROTEIN, ... | | Authors: | Chen, J.C.-H, Krucinski, J, Miercke, L.J.W, Finer-Moore, J.S, Tang, A.H, Leavitt, A.D, Stroud, R.M. | | Deposit date: | 2000-05-03 | | Release date: | 2000-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
3ZCF
 
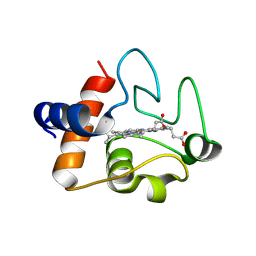 | | Structure of recombinant human cytochrome c | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Rajagopal, B.S, Worrall, J.A.R, Hough, M.A. | | Deposit date: | 2012-11-20 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Hydrogen Peroxide Induced Radical Behaviour in Human Cytochrome C Phospholipid Complexes: Implications for the Enhanced Pro-Apoptotic Activity of the G41S Mutant
Biochem.J., 456, 2013
|
|
2IWN
 
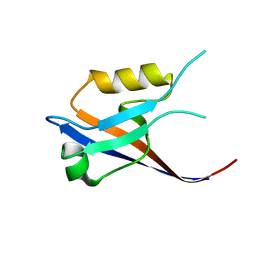 | | 3rd PDZ domain of Multiple PDZ Domain Protein MPDZ | | Descriptor: | MULTIPLE PDZ DOMAIN PROTEIN | | Authors: | Elkins, J.M, Gileadi, C, Savitsky, P, Berridge, G, Smee, C.E.A, Pike, A.C.W, Papagrigoriou, E, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Doyle, D.A. | | Deposit date: | 2006-07-03 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of Pick1 and Other Pdz Domains Obtained with the Help of Self-Binding C-Terminal Extension.
Protein Sci., 16, 2007
|
|
1GYO
 
 | | Crystal structure of the di-tetraheme cytochrome c3 from Desulfovibrio gigas at 1.2 Angstrom resolution | | Descriptor: | CYTOCHROME C3, A DIMERIC CLASS III C-TYPE CYTOCHROME, GLYCEROL, ... | | Authors: | Aragao, D, Frazao, C, Sieker, L, Sheldrick, G.M, Legall, J, Carrondo, M.A. | | Deposit date: | 2002-04-29 | | Release date: | 2002-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Dimeric Cytochrome C3 from Desulfovibrio Gigas at 1.2 A Resolution
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1YIC
 
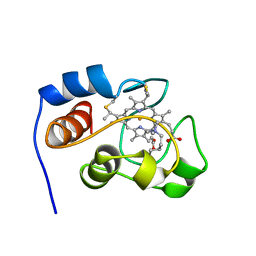 | | THE OXIDIZED SACCHAROMYCES CEREVISIAE ISO-1-CYTOCHROME C, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C, ISO-1, HEME C | | Authors: | Banci, L, Bertini, I, Bren, K.L, Gray, H.B, Sompornpisut, P, Turano, P. | | Deposit date: | 1997-02-18 | | Release date: | 1997-07-23 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of oxidized Saccharomyces cerevisiae iso-1-cytochrome c.
Biochemistry, 36, 1997
|
|
4JXT
 
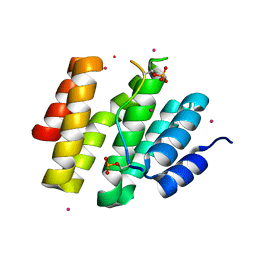 | | CID of human RPRD1A in complex with a phosphorylated peptide from RPB1-CTD | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Regulation of nuclear pre-mRNA domain-containing protein 1A, UNKNOWN ATOM OR ION | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Guo, X, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-28 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1CPF
 
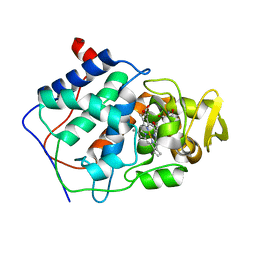 | | A CATION BINDING MOTIF STABILIZES THE COMPOUND I RADICAL OF CYTOCHROME C PEROXIDASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Miller, M.A, Han, G.W, Kraut, J. | | Deposit date: | 1994-08-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cation binding motif stabilizes the compound I radical of cytochrome c peroxidase.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1ZTG
 
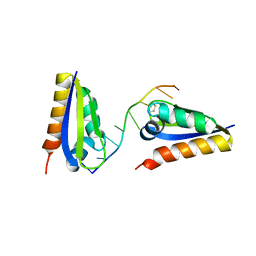 | | human alpha polyC binding protein KH1 | | Descriptor: | 5'-D(P*CP*CP*CP*TP*CP*CP*CP*T)-3', POLY(RC)-BINDING PROTEIN 1 | | Authors: | Sidiqi, M, Wilce, J.A, Barker, A, Schmidgerger, J, Leedman, P.J, Wilce, M.C.J. | | Deposit date: | 2005-05-27 | | Release date: | 2006-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Contribution of the first K-homology domain of poly(C)-binding protein 1 to its affinity and specificity for C-rich oligonucleotides
Nucleic Acids Res., 40, 2012
|
|
2MJF
 
 | | Solution structure of the complex between the yeast Rsa1 and Hit1 proteins | | Descriptor: | Protein HIT1, Ribosome assembly 1 protein | | Authors: | Quinternet, M, Roth, B, Back, R, Jacquemin, C, Manival, X. | | Deposit date: | 2014-01-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein Hit1, a novel box C/D snoRNP assembly factor, controls cellular concentration of the scaffolding protein Rsa1 by direct interaction.
Nucleic Acids Res., 42, 2014
|
|
1CPD
 
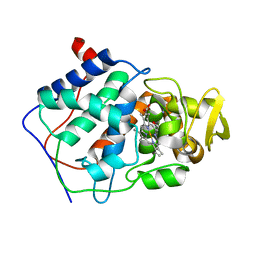 | |
1ZAP
 
 | | SECRETED ASPARTIC PROTEASE FROM C. ALBICANS | | Descriptor: | N-ethyl-N-[(4-methylpiperazin-1-yl)carbonyl]-D-phenylalanyl-N-[(1S,2S,4R)-4-(butylcarbamoyl)-1-(cyclohexylmethyl)-2-hyd roxy-5-methylhexyl]-L-norleucinamide, SECRETED ASPARTIC PROTEINASE, ZINC ION | | Authors: | Abad-Zapatero, C, Muchmore, S.W. | | Deposit date: | 1996-01-16 | | Release date: | 1997-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a secreted aspartic protease from C. albicans complexed with a potent inhibitor: implications for the design of antifungal agents.
Protein Sci., 5, 1996
|
|
1E08
 
 | | Structural model of the [Fe]-Hydrogenase/cytochrome c553 complex combining NMR and soft-docking | | Descriptor: | 1,3-PROPANEDITHIOL, CARBON MONOXIDE, CYANIDE ION, ... | | Authors: | Morelli, X, Czjzek, M, Hatchikian, C.E, Bornet, O, Fontecilla-Camps, J.C, Palma, N.P, Moura, J.J.G, Guerlesquin, F. | | Deposit date: | 2000-03-13 | | Release date: | 2000-08-25 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | Structural Model of the Fe-Hydrogenase/Cytochrome C553 Complex Combining Transverse Relaxation-Optimized Spectroscopy Experiments and Soft Docking Calculations.
J.Biol.Chem., 275, 2000
|
|
4RSZ
 
 | |
