3CGC
 
 | |
3DFZ
 
 | | SirC, precorrin-2 dehydrogenase | | Descriptor: | GLYCEROL, Precorrin-2 dehydrogenase, SULFATE ION | | Authors: | Schubert, H.L, Hill, C.P, Warren, M.J. | | Deposit date: | 2008-06-12 | | Release date: | 2008-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of SirC from Bacillus megaterium: a metal-binding precorrin-2 dehydrogenase
Biochem.J., 415, 2008
|
|
1C9E
 
 | | STRUCTURE OF FERROCHELATASE WITH COPPER(II) N-METHYLMESOPORPHYRIN COMPLEX BOUND AT THE ACTIVE SITE | | Descriptor: | MAGNESIUM ION, N-METHYLMESOPORPHYRIN CONTAINING COPPER, PROTOHEME FERROLYASE | | Authors: | Lecerof, D, Fodje, M.N, Hansson, A, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 1999-08-02 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic basis of porphyrin metallation by ferrochelatase.
J.Mol.Biol., 297, 2000
|
|
7BBN
 
 | |
7BN3
 
 | |
7BFC
 
 | |
7BFD
 
 | |
1E5L
 
 | | Apo saccharopine reductase from Magnaporthe grisea | | Descriptor: | SACCHAROPINE REDUCTASE | | Authors: | Johansson, E, Steffens, J.J, Lindqvist, Y, Schneider, G. | | Deposit date: | 2000-07-27 | | Release date: | 2000-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Saccharopine Reductase from Magnaporthe Grisea, an Enzyme of the Alpha-Aminoadipate Pathway of Lysine Biosynthesis
Structure, 8, 2000
|
|
7NDV
 
 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001888. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[4-(trifluoromethyl)phenoxy]piperidine, Acetylcholine-binding protein, ... | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2021-02-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of fragments inducing conformational effects in dynamic proteins using a second-harmonic generation biosensor
RSC Advances, 11, 2021
|
|
7NDP
 
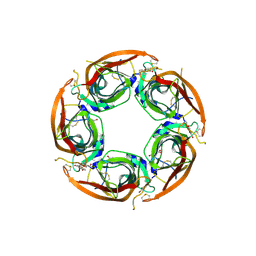 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001856. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-bromanylspiro[3~{H}-chromene-2,4'-piperidine]-4-one, ... | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2021-02-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of fragments inducing conformational effects in dynamic proteins using a second-harmonic generation biosensor.
Rsc Adv, 11, 2021
|
|
1D7D
 
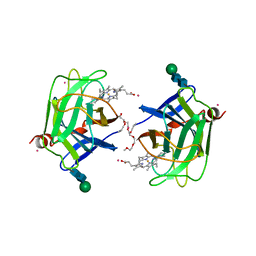 | | CYTOCHROME DOMAIN OF CELLOBIOSE DEHYDROGENASE, HP3 FRAGMENT, PH 7.5 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CADMIUM ION, CELLOBIOSE DEHYDROGENASE, ... | | Authors: | Hallberg, B.M, Bergfors, T, Backbro, K, Divne, C. | | Deposit date: | 1999-10-16 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new scaffold for binding haem in the cytochrome domain of the extracellular flavocytochrome cellobiose dehydrogenase.
Structure Fold.Des., 8, 2000
|
|
1G8P
 
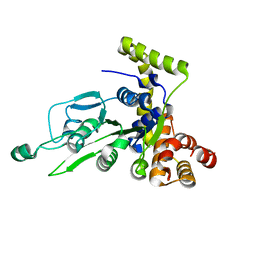 | | CRYSTAL STRUCTURE OF BCHI SUBUNIT OF MAGNESIUM CHELATASE | | Descriptor: | MAGNESIUM-CHELATASE 38 KDA SUBUNIT | | Authors: | Fodje, M.N, Hansson, A, Hansson, M, Olsen, J.G, Gough, S, Willows, R.D, Al-Karadaghi, S. | | Deposit date: | 2000-11-20 | | Release date: | 2001-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interplay between an AAA module and an integrin I domain may regulate the function of magnesium chelatase.
J.Mol.Biol., 311, 2001
|
|
1F77
 
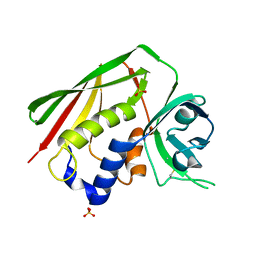 | | STAPHYLOCOCCAL ENTEROTOXIN H DETERMINED TO 2.4 A RESOLUTION | | Descriptor: | ENTEROTOXIN H, SULFATE ION | | Authors: | Hakansson, M, Petersson, K, Nilsson, H, Forsberg, G, Bjork, P. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of staphylococcal enterotoxin H: implications for binding properties to MHC class II and TcR molecules.
J.Mol.Biol., 302, 2000
|
|
6XWE
 
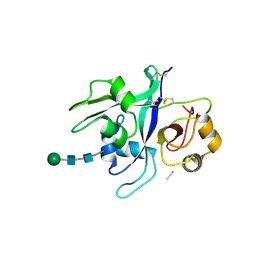 | | Crystal structure of LYK3 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETONITRILE, LysM domain receptor-like kinase 3, ... | | Authors: | Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-01-23 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity.
Science, 369, 2020
|
|
6YK4
 
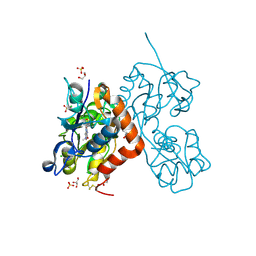 | | Structure of the AMPA receptor GluA2o ligand-binding domain (S1S2J) in complex with the compound ( S) - 1- [2'-Amino-2'-carboxyethyl]-6-methyl-5 ,7- dihydropyrrolo[3,4-d]pyrimidin-2,4(1H,3H)-dione at resolution 1.00A | | Descriptor: | (2~{S})-2-azanyl-3-[6-methyl-2,4-bis(oxidanylidene)-5,7-dihydropyrrolo[3,4-d]pyrimidin-1-yl]propanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2020-04-05 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.999 Å) | | Cite: | Ionotropic Glutamate Receptor GluA2 in Complex with Bicyclic Pyrimidinedione-Based Compounds: When Small Compound Modifications Have Distinct Effects on Binding Interactions.
Acs Chem Neurosci, 11, 2020
|
|
6YF7
 
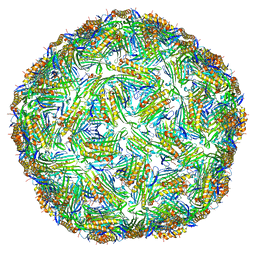 | | Virus-like particle of bacteriophage AC | | Descriptor: | Coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
9GK1
 
 | |
6YK3
 
 | | Structure of the AMPA receptor GluA2o ligand-binding domain (S1S2J) in complex with the compound ( S) - 1- [2'-Amino-2'-carboxyethyl]-5 ,7- dihydropyrrolo[3,4-d]pyrimidin-2,4(1H,3H)-dione at resolution 1.20A | | Descriptor: | (S)-1-[2'-Amino-2'-carboxyethyl]-5,7-dihydropyrrolo[3,4-d]pyrimidin-2,4(1H,3H)-dione, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2020-04-05 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ionotropic Glutamate Receptor GluA2 in Complex with Bicyclic Pyrimidinedione-Based Compounds: When Small Compound Modifications Have Distinct Effects on Binding Interactions.
Acs Chem Neurosci, 11, 2020
|
|
6YK5
 
 | | Structure of the AMPA receptor GluA2o ligand-binding domain (S1S2J) in complex with the compound (S)-1-(2'-Amino-2'-carboxyethyl)-5,7-dihydrofuro[3,4-d]- pyrimidine-2,4(1H,3H)-dione at resolution 1.15A | | Descriptor: | (S)-1-(2'-Amino-2'-carboxyethyl)-5,7-dihydrofuro[3,4-d]-pyrimidine-2,4(1H,3H)-dione, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2020-04-05 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ionotropic Glutamate Receptor GluA2 in Complex with Bicyclic Pyrimidinedione-Based Compounds: When Small Compound Modifications Have Distinct Effects on Binding Interactions.
Acs Chem Neurosci, 11, 2020
|
|
6YK6
 
 | | Structure of the AMPA receptor GluA2o ligand-binding domain (S1S2J) in complex with the compound (S)-1-(2'-Amino-2'-carboxyethyl)furo[3,4-d]pyrimidin-2,4-dione at resolution 1.47A | | Descriptor: | (S)-1-(2'-Amino-2'-carboxyethyl)furo[3,4-d]pyrimidin-2,4-dione, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2020-04-05 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.469 Å) | | Cite: | Ionotropic Glutamate Receptor GluA2 in Complex with Bicyclic Pyrimidinedione-Based Compounds: When Small Compound Modifications Have Distinct Effects on Binding Interactions.
Acs Chem Neurosci, 11, 2020
|
|
6HMY
 
 | |
6QLS
 
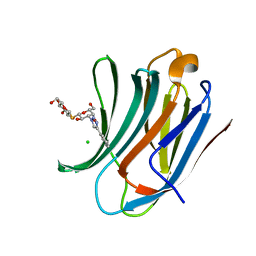 | | Galectin-3C in complex with fluoroaryltriazole monothiogalactoside derivative 6 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-(4-phenyl-1,2,3-triazol-1-yl)oxan-2-yl]sulfanyl-oxane-3,4,5-triol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.047 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
4CNU
 
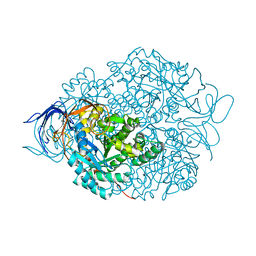 | |
4CML
 
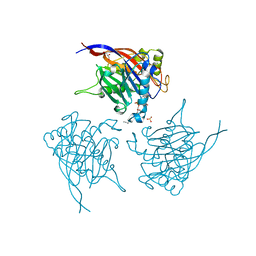 | | Crystal Structure of INPP5B in complex with Phosphatidylinositol 3,4- bisphosphate | | Descriptor: | 1,2-dioctanoyl phosphatidyl epi-inositol (3,4)-bisphosphate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Flodin, S, Graslund, S, Karlberg, T, Moche, M, Nyman, T, Schuler, H, Silvander, C, Thorsell, A.G, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Phosphoinositide Substrate Recognition, Catalysis, and Membrane Interactions in Human Inositol Polyphosphate 5-Phosphatases.
Structure, 22, 2014
|
|
4CKU
 
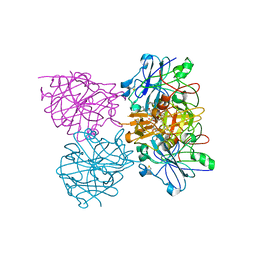 | | Three dimensional structure of plasmepsin II in complex with hydroxyethylamine-based inhibitor | | Descriptor: | 5-[1,1-bis(oxidanylidene)-1,2-thiazinan-2-yl]-N3-[(2S,3R)-4-[2-(3-methoxyphenyl)propan-2-ylamino]-3-oxidanyl-1-phenyl-butan-2-yl]-N1,N1-dipropyl-benzene-1,3-dicarboxamide, PLASMEPSIN-2 | | Authors: | Tars, K, Leitans, J, Jaudzems, K. | | Deposit date: | 2014-01-08 | | Release date: | 2014-06-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Plasmepsin Inhibitory Activity and Structure-Guided Optimization of a Potent Hydroxyethylamine-Based Antimalarial Hit.
Acs Med.Chem.Lett., 5, 2014
|
|
