7CB0
 
 | |
7CB5
 
 | |
7COH
 
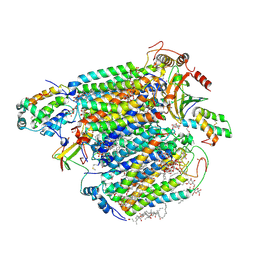 | | Dimeric Form of Bovine Heart Cytochrome c Oxidase in the Fully Oxidized State | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, 1,2-ETHANEDIOL, ... | | Authors: | Shinzawa-Itoh, K, Muramoto, K. | | Deposit date: | 2020-08-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3-A Resolution structure of bovine cytochrome c oxidase suggests a dimerization mechanism
Biochim.Biophys.Acta, 2021
|
|
1T34
 
 | | ROTATION MECHANISM FOR TRANSMEMBRANE SIGNALING BY THE ATRIAL NATRIURETIC PEPTIDE RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Atrial natriuretic peptide factor, Atrial natriuretic peptide receptor A, ... | | Authors: | Ogawa, H, Qiu, Y, Ogata, C.M, Misono, K.S. | | Deposit date: | 2004-04-23 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of hormone-bound atrial natriuretic peptide receptor extracellular domain: rotation mechanism for transmembrane signal transduction
J.Biol.Chem., 279, 2004
|
|
7CB6
 
 | |
7CGS
 
 | |
1UD2
 
 | | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) | | Descriptor: | GLYCEROL, SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1TVR
 
 | | HIV-1 RT/9-CL TIBO | | Descriptor: | 4-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Das, K, Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 1996-04-16 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant.
J.Mol.Biol., 264, 1996
|
|
1UDR
 
 | |
1UGE
 
 | |
1TVW
 
 | |
1TSW
 
 | | THYMIDYLATE SYNTHASE R179A MUTANT | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-12-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contribution of a salt bridge to binding affinity and dUMP orientation to catalytic rate: mutation of a substrate-binding arginine in thymidylate synthase.
Protein Eng., 9, 1996
|
|
1TSZ
 
 | | THYMIDYLATE SYNTHASE R179K MUTANT | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-12-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Contribution of a salt bridge to binding affinity and dUMP orientation to catalytic rate: mutation of a substrate-binding arginine in thymidylate synthase.
Protein Eng., 9, 1996
|
|
7CYN
 
 | | Cryo-EM structure of human TLR7 in complex with UNC93B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein unc-93 homolog B1, ... | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-06 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of Toll-like receptors in complex with UNC93B1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1TSL
 
 | |
1TSM
 
 | |
7D7N
 
 | | Cryo-EM structure of human ABCB6 transporter | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial | | Authors: | Wang, C, Cao, C, Wang, N, Wang, X, Zhang, X.C. | | Deposit date: | 2020-10-05 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-electron microscopy structure of human ABCB6 transporter.
Protein Sci., 29, 2020
|
|
1TMO
 
 | | TRIMETHYLAMINE N-OXIDE REDUCTASE FROM SHEWANELLA MASSILIA | | Descriptor: | GUANYLATE-O'-PHOSPHORIC ACID MONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,5,6,7,8A,9,10,10A-OCTAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL) ESTER, MOLYBDENUM (IV)OXIDE, TRIMETHYLAMINE N-OXIDE REDUCTASE | | Authors: | Czjzek, M, Dos Santos, J.P, Giordano, G, Mejean, V. | | Deposit date: | 1998-08-03 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of oxidized trimethylamine N-oxide reductase from Shewanella massilia at 2.5 A resolution.
J.Mol.Biol., 284, 1998
|
|
7D6B
 
 | | Crystal structure of Oryza sativa Os4BGlu18 monolignol beta-glucosidase with delta-gluconolactone | | Descriptor: | Beta-glucosidase 18, D-glucono-1,5-lactone, GLYCEROL, ... | | Authors: | Baiya, S, Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2020-09-29 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of rice Os4BGlu18 monolignol beta-glucosidase.
Plos One, 16, 2021
|
|
1TPU
 
 | | S96P CHANGE IS A SECOND-SITE SUPPRESSOR FOR H95N SLUGGISH MUTANT TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Stock, A.M, Narayana, N, Xuong, Ng.H, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-11-07 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the H95N lesion by the secondary S96P mutation in triosephosphate isomerase.
Biochemistry, 35, 1996
|
|
1TPV
 
 | | S96P CHANGE IS A SECOND-SITE SUPPRESSOR FOR H95N SLUGGISH MUTANT TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Stock, A.M, Narayana, N, Xuong, Ng.H, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-11-07 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the H95N lesion by the secondary S96P mutation in triosephosphate isomerase.
Biochemistry, 35, 1996
|
|
7D6A
 
 | | Crystal structure of Oryza sativa Os4BGlu18 monolignol beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 18, GLYCEROL, ... | | Authors: | Baiya, S, Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2020-09-29 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of rice Os4BGlu18 monolignol beta-glucosidase.
Plos One, 16, 2021
|
|
7DFM
 
 | |
7D94
 
 | | Crystal Structure of the Na+,K+-ATPase in the E2P state with bound one Mg2+ and one Rb+ in the presence of bufalin | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Ogawa, H, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-12 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1TPB
 
 | | OFFSET OF A CATALYTIC LESION BY A BOUND WATER SOLUBLE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-03 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the E165D lesion by the secondary S96P mutation in triosephosphate isomerase depends on the positions of active site water molecules.
Biochemistry, 34, 1995
|
|
