1TXE
 
 | | Solution structure of the active-centre mutant Ile14Ala of the histidine-containing phosphocarrier protein (HPr) from Staphylococcus carnosus | | Descriptor: | Phosphocarrier protein HPr | | Authors: | Moeglich, A, Koch, B, Hengstenberg, W, Brunner, E, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-07-04 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the active-centre mutant I14A of the histidine-containing phosphocarrier protein from Staphylococcus carnosus
Eur.J.Biochem., 271, 2004
|
|
1TXF
 
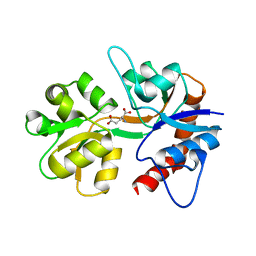 | |
1TXG
 
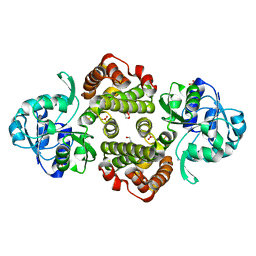 | | Structure of glycerol-3-phosphate dehydrogenase from Archaeoglobus fulgidus | | Descriptor: | AMMONIUM ION, GLYCEROL, SULFATE ION, ... | | Authors: | Sakasegawa, S, Hagemeier, C.H, Thauer, R.K, Essen, L.O, Shima, S. | | Deposit date: | 2004-07-05 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional analysis of the gpsA gene product of Archaeoglobus fulgidus: A glycerol-3-phosphate dehydrogenase with an unusual NADP+ preference
Protein Sci., 13, 2004
|
|
1TXI
 
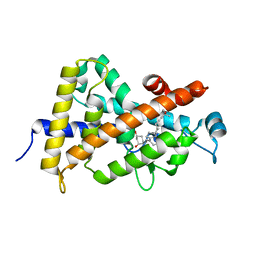 | | Crystal structure of the vdr ligand binding domain complexed to TX522 | | Descriptor: | (1R,3R)-5-((Z)-2-((1R,7AS)-HEXAHYDRO-1-((S)-6-HYDROXY-6-METHYLHEPT-4-YN-2-YL)-7A-METHYL-1H-INDEN-4(7AH)-YLIDENE)ETHYLIDENE)CYCLOHEXANE-1,3-DIOL, Vitamin D receptor | | Authors: | Moras, D, Rochel, N. | | Deposit date: | 2004-07-05 | | Release date: | 2005-05-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Superagonistic Action of 14-epi-Analogs of 1,25-Dihydroxyvitamin D Explained by Vitamin D Receptor-Coactivator Interaction
Mol.Pharmacol., 67, 2005
|
|
1TXJ
 
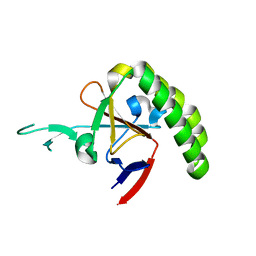 | | Crystal structure of translationally controlled tumour-associated protein (TCTP) from Plasmodium knowlesi | | Descriptor: | translationally controlled tumour-associated protein (TCTP) from Plasmodium knowlesi, PKN_PFE0545c | | Authors: | Walker, J.R, Vedadi, M, Sharma, S, Houston, S, Lew, J, Amani, M, Wasney, G, Skarina, T, Bray, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-07-05 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
1TXK
 
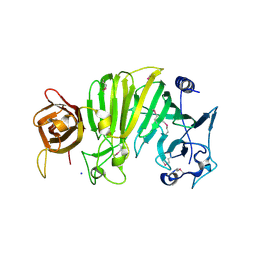 | | Crystal structure of Escherichia coli OpgG | | Descriptor: | Glucans biosynthesis protein G, SODIUM ION | | Authors: | Hanoulle, X, Rollet, E, Clantin, B, Landrieu, I, Odberg-Ferragut, C, Lippens, G, Bohin, J.P, Villeret, V. | | Deposit date: | 2004-07-05 | | Release date: | 2004-09-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis of Escherichia coli OpgG, a Protein Required for the Biosynthesis of Osmoregulated Periplasmic Glucans.
J.Mol.Biol., 342, 2004
|
|
1TXL
 
 | |
1TXM
 
 | | SCORPION TOXIN (MAUROTOXIN) FROM SCORPIO MAURUS, NMR, 35 STRUCTURES | | Descriptor: | MAUROTOXIN | | Authors: | Blanc, E, Sabatier, J.-M, Kharrat, R, Meunier, S, El Ayeb, M, Van Rietschoten, J, Darbon, H. | | Deposit date: | 1996-11-19 | | Release date: | 1997-06-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of maurotoxin, a scorpion toxin from Scorpio maurus, with high affinity for voltage-gated potassium channels.
Proteins, 29, 1997
|
|
1TXN
 
 | |
1TXO
 
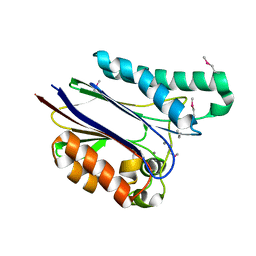 | | Crystal structure of the Mycobacterium tuberculosis serine/threonine phosphatase PstP/Ppp at 1.95 A. | | Descriptor: | MANGANESE (II) ION, Putative Bacterial Enzyme | | Authors: | Pullen, K.E, Ng, H.L, Sung, P.Y, Good, M.C, Smith, S.M, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-07-05 | | Release date: | 2004-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An Alternate Conformation and a Third Metal in PstP/Ppp, the M. tuberculosis PP2C-Family Ser/Thr Protein Phosphatase.
Structure, 12, 2004
|
|
1TXP
 
 | |
1TXQ
 
 | |
1TXR
 
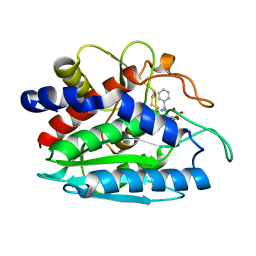 | | X-ray crystal structure of bestatin bound to AAP | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Bacterial leucyl aminopeptidase, ZINC ION | | Authors: | Stamper, C.C, Holz, R.C, Ringe, D, Petsko, G.A. | | Deposit date: | 2004-07-06 | | Release date: | 2004-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Spectroscopic and X-ray Crystallographic Characterization of Bestatin Bound to the Aminopeptidase from Aeromonas (Vibrio) proteolytica.
Biochemistry, 43, 2004
|
|
1TXS
 
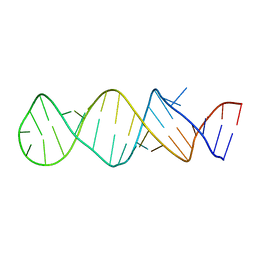 | | STEM-LOOP D OF THE CLOVERLEAF DOMAIN OF ENTEROVIRAL 5'UTR RNA | | Descriptor: | Enteroviral 5'-UTR | | Authors: | Du, Z, Yu, J, Ulyanov, N.B, Andino, R, James, T.L. | | Deposit date: | 2004-07-06 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Consensus Stem-Loop D RNA Domain that Plays Important Roles in Regulating Translation and Replication in Enteroviruses and Rhinoviruses
Biochemistry, 43, 2004
|
|
1TXT
 
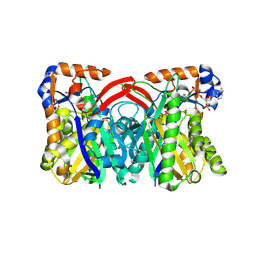 | | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase | | Descriptor: | 3-hydroxy-3-methylglutaryl-CoA synthase, ACETOACETYL-COENZYME A | | Authors: | Campobasso, N, Patel, M, Wilding, I.E, Kallender, H, Rosenberg, M, Gwynn, M. | | Deposit date: | 2004-07-06 | | Release date: | 2004-08-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism
J.Biol.Chem., 279, 2004
|
|
1TXU
 
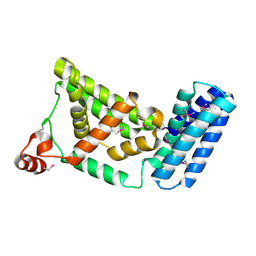 | | Crystal Structure of the Vps9 Domain of Rabex-5 | | Descriptor: | MAGNESIUM ION, Rab5 GDP/GTP exchange factor | | Authors: | Delprato, A, Merithew, E, Lambright, D.G. | | Deposit date: | 2004-07-06 | | Release date: | 2004-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure, exchange determinants, and family-wide rab specificity of the tandem helical bundle and Vps9 domains of Rabex-5
Cell(Cambridge,Mass.), 118, 2004
|
|
1TXX
 
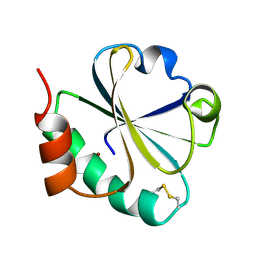 | |
1TXY
 
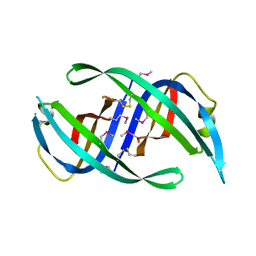 | | E. coli PriB | | Descriptor: | Primosomal replication protein n | | Authors: | Keck, J.L, Lopper, M, Holton, J.M. | | Deposit date: | 2004-07-06 | | Release date: | 2004-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PriB, a component of the Escherichia coli replication restart primosome
Structure, 12, 2004
|
|
1TXZ
 
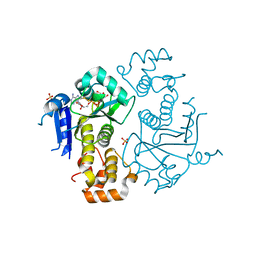 | | Crystal structure of yeast ymx7, an ADP-ribose-1''-monophosphatase, complexed with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Hypothetical 32.1 kDa protein in ADH3-RCA1 intergenic region, ... | | Authors: | Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-06 | | Release date: | 2004-11-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and mechanism of ADP-ribose-1''-monophosphatase (Appr-1''-pase), a ubiquitous cellular processing enzyme.
Protein Sci., 14, 2005
|
|
1TY0
 
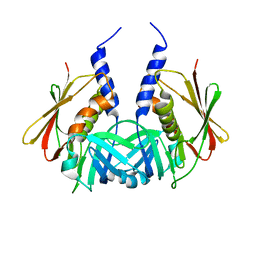 | | Crystal structure of the streptococcal pyrogenic exotoxin J (SPE-J) | | Descriptor: | putative exotoxin (superantigen) | | Authors: | Baker, H.M, Proft, T, Webb, P.D, Arcus, V.L, Fraser, J.D, Baker, E.N. | | Deposit date: | 2004-07-06 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic and mutational data show that the streptococcal pyrogenic exotoxin j can use a common binding surface for T-cell receptor binding and dimerization
J.Biol.Chem., 279, 2004
|
|
1TY2
 
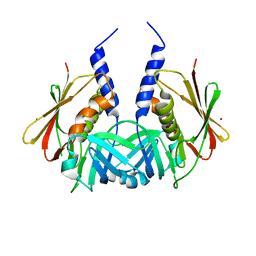 | | Crystal structure of the streptococcal pyrogenic exotoxin J (SPE-J) | | Descriptor: | ZINC ION, putative exotoxin (superantigen) | | Authors: | Baker, H.M, Proft, T, Webb, P.D, Arcus, V.L, Fraser, J.D, Baker, E.N. | | Deposit date: | 2004-07-07 | | Release date: | 2004-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and mutational data show that the streptococcal pyrogenic exotoxin j can use a common binding surface for T-cell receptor binding and dimerization
J.Biol.Chem., 279, 2004
|
|
1TY4
 
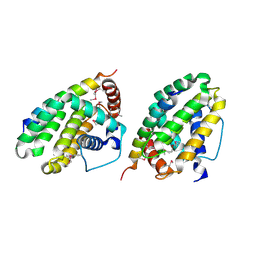 | | Crystal structure of a CED-9/EGL-1 complex | | Descriptor: | Apoptosis regulator ced-9, EGg Laying defective EGL-1, programmed cell death activator | | Authors: | Yan, N, Gu, L, Kokel, D, Xue, D, Shi, Y. | | Deposit date: | 2004-07-07 | | Release date: | 2004-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural, Biochemical, and Functional Analyses of CED-9 Recognition by the Proapoptotic Proteins EGL-1 and CED-4
Mol.Cell, 15, 2004
|
|
1TY8
 
 | | Crystal structure of yeast ymx7, an ADP-ribose-1''-monophosphatase, complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Hypothetical 32.1 kDa protein in ADH3-RCA1 intergenic region, ... | | Authors: | Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-07 | | Release date: | 2004-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF YEAST HYPOTHETICAL PROTEIN YMX7
To be Published
|
|
1TY9
 
 | | X-RAY CRYSTAL STRUCTURE OF PHZG FROM PSEUDOMONAS FLUORESCENS | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phenazine biosynthesis protein phzG, SULFATE ION | | Authors: | Parsons, J.F, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2004-07-07 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the phenazine biosynthesis enzyme PhzG.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TYA
 
 | |
