6YF0
 
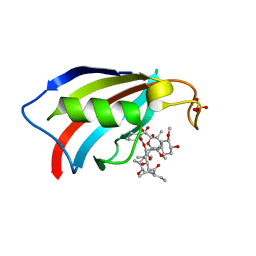 | | FKBP12 in complex with the BMP potentiator compound 9 at 1.55 A resolution | | Descriptor: | 18-HYDROXYASCOMYCIN, Peptidyl-prolyl cis-trans isomerase FKBP1A, SULFATE ION | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
6MRD
 
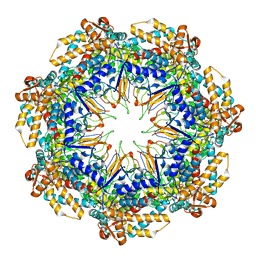 | | ADP-bound human mitochondrial Hsp60-Hsp10 half-football complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Gomez-Llorente, Y, Jebara, F, Patra, M, Malik, R, Nissemblat, S, Azem, A, Hirsch, J.A, Ubarretxena-Belandia, I. | | Deposit date: | 2018-10-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for active single and double ring complexes in human mitochondrial Hsp60-Hsp10 chaperonin.
Nat Commun, 11, 2020
|
|
1F40
 
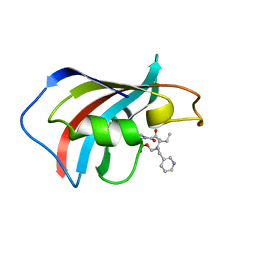 | | SOLUTION STRUCTURE OF FKBP12 COMPLEXED WITH GPI-1046, A NEUROTROPHIC LIGAND | | Descriptor: | (2S)-[3-PYRIDYL-1-PROPYL]-1-[3,3-DIMETHYL-1,2-DIOXOPENTYL]-2-PYRROLIDINECARBOXYLATE, FK506 BINDING PROTEIN (FKBP12) | | Authors: | Sich, C, Improta, S, Cowley, D.J, Guenet, C, Merly, J.P, Teufel, M, Saudek, V. | | Deposit date: | 2000-06-07 | | Release date: | 2000-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a neurotrophic ligand bound to FKBP12 and its effects on protein dynamics.
Eur.J.Biochem., 267, 2000
|
|
6MQ6
 
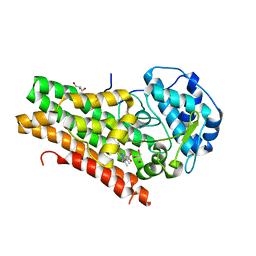 | | Mapping the binding trajectory of a suicide inhibitor in human indoleamine 2,3-dioxygenase 1 | | Descriptor: | (2R)-N-(4-chlorophenyl)-2-[cis-4-(6-fluoroquinolin-4-yl)cyclohexyl]propanamide, GLYCEROL, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Pham, K.N, Yeh, S.R. | | Deposit date: | 2018-10-09 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Mapping the Binding Trajectory of a Suicide Inhibitor in Human Indoleamine 2,3-Dioxygenase 1.
J. Am. Chem. Soc., 140, 2018
|
|
2YRW
 
 | | Crystal structure of GAR synthetase from Geobacillus kaustophilus | | Descriptor: | PHOSPHATE ION, Phosphoribosylglycinamide synthetase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
2YS6
 
 | | Crystal structure of GAR synthetase from Geobacillus kaustophilus | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCINE, Phosphoribosylglycinamide synthetase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
2YS7
 
 | | Crystal structure of GAR synthetase from Geobacillus kaustophilus | | Descriptor: | Phosphoribosylglycinamide synthetase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
2Z02
 
 | | Crystal structure of phosphoribosylaminoimidazolesuccinocarboxamide synthase wit ATP from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, Phosphoribosylaminoimidazole-succinocarboxamide synthase, ... | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of phosphoribosylaminoimidazolesuccinocarboxamide synthase from Methanocaldococcus jannaschii
To be Published
|
|
1KJX
 
 | | IMP Complex of E. Coli Adenylosuccinate Synthetase | | Descriptor: | Adenylosuccinate Synthetase, INOSINIC ACID | | Authors: | Hou, Z, Wang, W, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | IMP Alone Organizes the Active Site of Adenylosuccinate Synthetase from Escherichia coli.
J.Biol.Chem., 277, 2002
|
|
2AEE
 
 | | Crystal structure of Orotate phosphoribosyltransferase from Streptococcus pyogenes | | Descriptor: | CHLORIDE ION, GLYCEROL, Orotate phosphoribosyltransferase, ... | | Authors: | Chang, C, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-22 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Orotate phosphoribosyltransferase from Streptococcus pyogenes
To be Published
|
|
2YWR
 
 | | Crystal structure of GAR transformylase from Aquifex aeolicus | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures and reaction mechanisms of the two related enzymes, PurN and PurU.
J.Biochem., 154, 2013
|
|
2YWX
 
 | | Crystal structure of phosphoribosylaminoimidazole carboxylase catalytic subunit from Methanocaldococcus jannaschii | | Descriptor: | Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of phosphoribosylaminoimidazole carboxylase catalytic subunit from Methanocaldococcus jannaschii
To be Published
|
|
2YX5
 
 | | Crystal Structure of Methanocaldococcus jannaschii PurS, One of the Subunits of Formylglycinamide Ribonucleotide Amidotransferase in the Purine Biosynthetic Pathway | | Descriptor: | UPF0062 protein MJ1593 | | Authors: | Kanagawa, M, Baba, S, Agari, Y, Chen, L.Q, Fu, Z.-Q, Chrzas, J, Wang, B.C, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-24 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Methanocaldococcus jannaschii PurS, One of the Subunits of Formylglycinamide Ribonucleotide Amidotransferase in the Purine Biosynthetic Pathway
To be Published
|
|
2AD5
 
 | | Mechanisms of feedback regulation and drug resistance of CTP synthetases: structure of the E. coli CTPS/CTP complex at 2.8-Angstrom resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Endrizzi, J.A, Kim, H, Anderson, P.M, Baldwin, E.P. | | Deposit date: | 2005-07-19 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanisms of Product Feedback Regulation and Drug Resistance in Cytidine Triphosphate Synthetases from the Structure of a CTP-Inhibited Complex(,).
Biochemistry, 44, 2005
|
|
1MC4
 
 | |
1MEZ
 
 | | Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with SAMP, GDP, SO4(2-), and Mg(2+) | | Descriptor: | 2-[9-(3,4-DIHYDROXY-5-PHOSPHONOOXYMETHYL-TETRAHYDRO-FURAN-2-YL)-9H-PURIN-6-YLAMINO]-SUCCINIC ACID, Adenylosuccinate Synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-08-09 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Feedback inhibition and product complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
2A1F
 
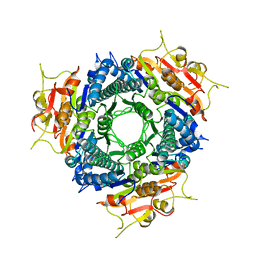 | |
1MF0
 
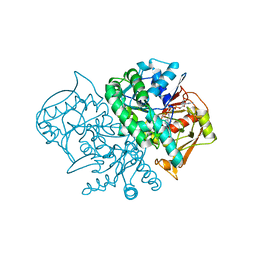 | | Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with AMP, GDP, HPO4(2-), and Mg(2+) | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylosuccinate Synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-08-09 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Feedback inhibition and product complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
1MF1
 
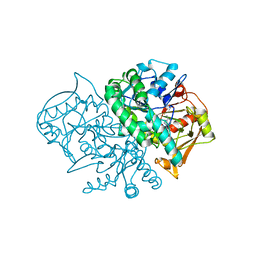 | | Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with AMP | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, Adenylosuccinate Synthetase | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-08-09 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Feedback inhibition and product complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
1GRC
 
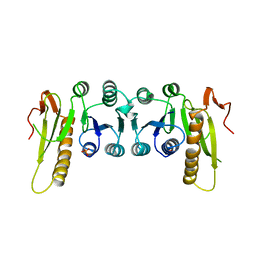 | |
6O3I
 
 | | Crystal Structure of Human IDO1 bound to navoximod (NLG-919) | | Descriptor: | Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, trans-4-{(1R)-2-[(5S)-6-fluoro-5H-imidazo[5,1-a]isoindol-5-yl]-1-hydroxyethyl}cyclohexan-1-ol | | Authors: | Harris, S.F, Oh, A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-17 | | Last modified: | 2019-08-07 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Clinical Candidate (1R,4r)-4-((R)-2-((S)-6-Fluoro-5H-imidazo[5,1-a]isoindol-5-yl)-1-hydroxyethyl)cyclohexan-1-ol (Navoximod), a Potent and Selective Inhibitor of Indoleamine 2,3-Dioxygenase 1.
J.Med.Chem., 62, 2019
|
|
2YZL
 
 | | Crystal structure of phosphoribosylaminoimidazole-succinocarboxamide synthase with ADP from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphoribosylaminoimidazole-succinocarboxamide synthase, SULFATE ION | | Authors: | Kanagawa, M, Baba, S, Fukui, K, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of phosphoribosylaminoimidazole-succinocarboxamide synthase with ADP from Methanocaldococcus jannaschii
To be Published
|
|
1FKH
 
 | | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS, AND THE X-RAY CRYSTAL STRUCTURES OF THEIR COMPLEXES WITH FKBP12 | | Descriptor: | 1-CYCLOHEXYL-3-PHENYL-1-PROPYL-1-(3,3-DIMETHYL-1,2-DIOXYPENTYL)-2-PIPERIDINE CARBOXYLATE, FK506 BINDING PROTEIN | | Authors: | Holt, D.A, Luengo, J.I, Yamashita, D.S, Oh, H.-J, Konialian, A.L, Yen, H.-K, Rozamus, L.W, Brandt, M, Bossard, M.J, Levy, M.A, Eggleston, D.S, Stout, T.J, Liang, J, Schultz, L.W, Clardy, J. | | Deposit date: | 1993-08-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS AND THE X-RAY CRYSTAL-STRUCTURES OF THEIR COMPLEXES WITH FKBP12.
J.Am.Chem.Soc., 115, 1993
|
|
2YWV
 
 | | Crystal structure of SAICAR synthetase from Geobacillus kaustophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylaminoimidazole succinocarboxamide synthetase, ... | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of SAICAR synthetase from Geobacillus kaustophilus
To be Published
|
|
6O55
 
 | |
