6JCX
 
 | |
8F1I
 
 | | SigN RNA polymerase early-melted intermediate bound to mismatch fragment dhsU36mm1 (-12T) | | Descriptor: | DNA (36-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Mueller, A.U, Chen, J, Darst, S.A. | | Deposit date: | 2022-11-05 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A general mechanism for transcription bubble nucleation in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F1J
 
 | | SigN RNA polymerase early-melted intermediate bound to mismatch DNA fragment dhsU36mm2 (-12A) | | Descriptor: | DNA (36-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Mueller, A.U, Chen, J, Darst, S.A. | | Deposit date: | 2022-11-05 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A general mechanism for transcription bubble nucleation in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F1K
 
 | | SigN RNA polymerase early-melted intermediate bound to full duplex DNA fragment dhsU36 (-12T) | | Descriptor: | DNA (36-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Mueller, A.U, Chen, J, Darst, S.A. | | Deposit date: | 2022-11-05 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A general mechanism for transcription bubble nucleation in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5A2V
 
 | | Crystal structure of mtPAP in Apo form | | Descriptor: | CHLORIDE ION, MITOCHONDRIAL PROTEIN | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5A2X
 
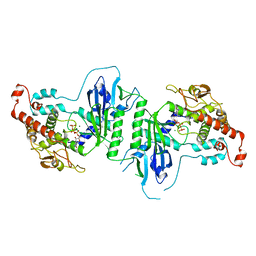 | | Crystal structure of mtPAP in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MITOCHONDRIAL PROTEIN | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5A2W
 
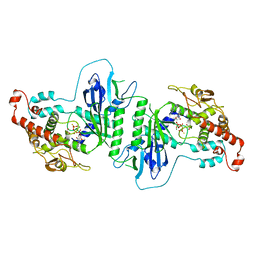 | | Crystal structure of mtPAP in complex with ATPgammaS | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, MITOCHONDRIAL PROTEIN, ... | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5A2Z
 
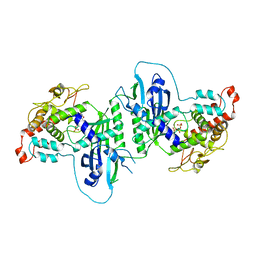 | | Crystal structure of mtPAP in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MITOCHONDRIAL PROTEIN | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5A2Y
 
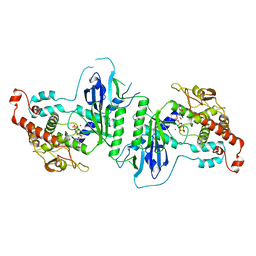 | | Crystal structure of mtPAP in complex with UTP | | Descriptor: | MAGNESIUM ION, MITOCHONDRIAL PROTEIN, URIDINE 5'-TRIPHOSPHATE | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5A30
 
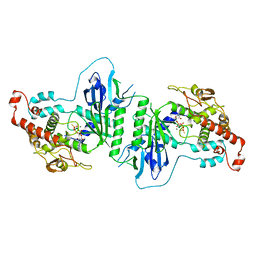 | | Crystal structure of mtPAP N472D mutant in complex with ATPgammaS | | Descriptor: | MAGNESIUM ION, MITOCHONDRIAL PROTEIN, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5JS2
 
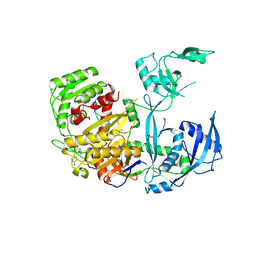 | | Human Argonaute-2 Bound to a Modified siRNA | | Descriptor: | MAGNESIUM ION, PHENOL, PHOSPHATE ION, ... | | Authors: | Schirle, N.T, MacRae, I.J. | | Deposit date: | 2016-05-07 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural Analysis of Human Argonaute-2 Bound to a Modified siRNA Guide.
J.Am.Chem.Soc., 138, 2016
|
|
5JS1
 
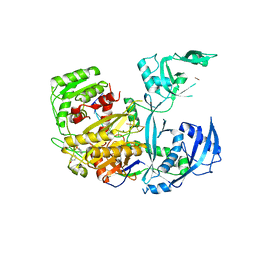 | | Human Argonaute2 Bound to an siRNA | | Descriptor: | MAGNESIUM ION, PHENOL, Protein argonaute-2, ... | | Authors: | Schirle, N.T, MacRae, I.J. | | Deposit date: | 2016-05-07 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural Analysis of Human Argonaute-2 Bound to a Modified siRNA Guide.
J.Am.Chem.Soc., 138, 2016
|
|
7EH0
 
 | |
8CYF
 
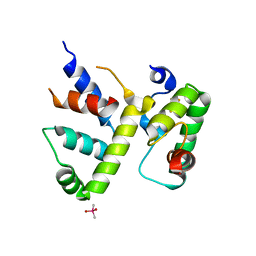 | | WhiB3 bound to SigmaAr4-RNAP Beta flap tip chimera and DNA | | Descriptor: | CACODYLATE ION, DNA (5'-D(*CP*AP*CP*CP*AP*CP*AP*AP*CP*CP*GP*AP*TP*TP*TP*T)-3'), DNA (5'-D(*GP*AP*AP*AP*AP*TP*CP*GP*GP*TP*TP*GP*TP*GP*GP*T)-3'), ... | | Authors: | Wan, T, Zhang, L.M. | | Deposit date: | 2022-05-23 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural basis of DNA binding by the WhiB-like transcription factor WhiB3 in Mycobacterium tuberculosis.
J.Biol.Chem., 299, 2023
|
|
7X76
 
 | |
7X75
 
 | |
7VPD
 
 | |
8Q3R
 
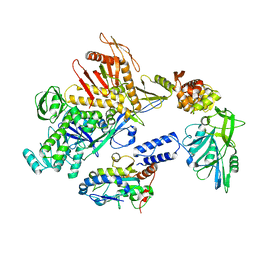 | | Cryo-EM structure of the DNA polymerase holoenzyme E9-A20-D4 of vaccinia virus | | Descriptor: | DNA polymerase, DNA polymerase processivity factor component OPG148, Uracil-DNA glycosylase | | Authors: | Burmeister, W.P, Ballandras-Colas, A, Boettcher, B, Grimm, C. | | Deposit date: | 2023-08-04 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and flexibility of the DNA polymerase holoenzyme of vaccinia virus.
Plos Pathog., 20, 2024
|
|
7CKL
 
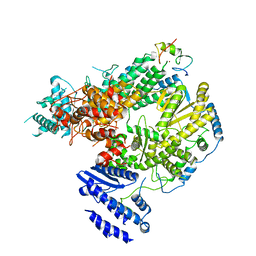 | | Structure of Lassa virus polymerase bound to Z matrix protein | | Descriptor: | MANGANESE (II) ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | Authors: | Xu, X, Peng, R, Peng, Q, Shi, Y. | | Deposit date: | 2020-07-17 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals
Nat Microbiol, 6, 2021
|
|
5HR7
 
 | | X-ray crystal structure of C118A RlmN from Escherichia coli with cross-linked in vitro transcribed tRNA | | Descriptor: | 5'-DEOXYADENOSINE, Dual-specificity RNA methyltransferase RlmN, IRON/SULFUR CLUSTER, ... | | Authors: | Schwalm, E.L, Grove, T.L, Booker, S.J, Boal, A.K. | | Deposit date: | 2016-01-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic capture of a radical S-adenosylmethionine enzyme in the act of modifying tRNA.
Science, 352, 2016
|
|
7X6V
 
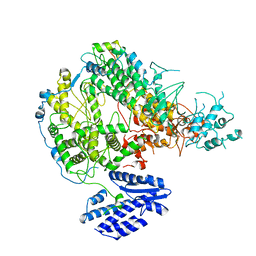 | |
7CKM
 
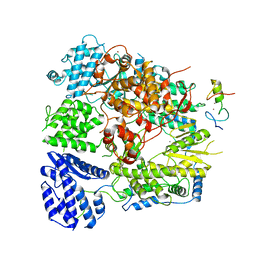 | | Structure of Machupo virus polymerase bound to Z matrix protein (monomeric complex) | | Descriptor: | MANGANESE (II) ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | Authors: | Xu, X, Peng, R, Peng, Q, Shi, Y. | | Deposit date: | 2020-07-17 | | Release date: | 2021-05-05 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals
Nat Microbiol, 6, 2021
|
|
8IZL
 
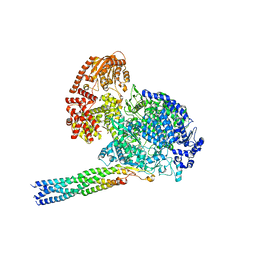 | |
4R71
 
 | | Structure of the Qbeta holoenzyme complex in the P1211 crystal form | | Descriptor: | 30S ribosomal protein S1, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Gytz, H, Seweryn, P, Kutlubaeva, Z, Chetverin, A.B, Brodersen, D.E, Knudsen, C.R. | | Deposit date: | 2014-08-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for RNA-genome recognition during bacteriophage Q beta replication.
Nucleic Acids Res., 43, 2015
|
|
8X01
 
 | |
