1GHD
 
 | | Crystal structure of the glutaryl-7-aminocephalosporanic acid acylase by mad phasing | | Descriptor: | GLUTARYL-7-AMINOCEPHALOSPORANIC ACID ACYLASE | | Authors: | Ding, Y, Jiang, W, Mao, X, He, H, Zhang, S, Tang, H, Bartlam, M, Ye, S, Jiang, F, Liu, Y, Zhao, G, Rao, Z. | | Deposit date: | 2000-12-07 | | Release date: | 2003-07-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Affinity alkylation of the Trp-B4 residue of the beta -subunit of the glutaryl 7-aminocephalosporanic acid acylase of Pseudomonas sp. 130.
J.Biol.Chem., 277, 2002
|
|
7F74
 
 | | Rv3094c in complex with FMN. | | Descriptor: | FLAVIN MONONUCLEOTIDE, Rv3094c | | Authors: | Wang, Z.X, Ouyang, S.Y. | | Deposit date: | 2021-06-27 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Omics analysis of Mycobacterium tuberculosis isolates uncovers Rv3094c, an ethionamide metabolism-associated gene.
Commun Biol, 6, 2023
|
|
1GE2
 
 | |
4YPH
 
 | | Crystal Structure of MutY bound to its anti-substrate with the disulfide cross-linker reduced | | Descriptor: | A/G-specific adenine glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), DNA (5'-D(TP*GP*TP*CP*CP*AP*CP*GP*TP*CP*T)-3'), ... | | Authors: | Wang, L, Lee, S, Verdine, G.L. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Basis for Avoidance of Promutagenic DNA Repair by MutY Adenine DNA Glycosylase.
J.Biol.Chem., 290, 2015
|
|
1FWH
 
 | |
3OID
 
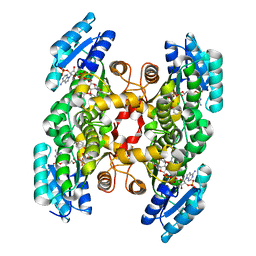 | | Crystal Structure of Enoyl-ACP Reductases III (FabL) from B. subtilis (complex with NADP and TCL) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRICLOSAN | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
7F5S
 
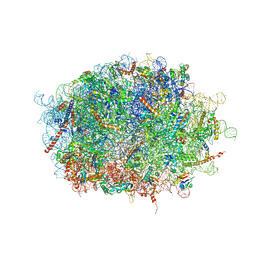 | | human delta-METTL18 60S ribosome | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Takahashi, M, Kashiwagi, K, Ito, T. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | METTL18-mediated histidine methylation of RPL3 modulates translation elongation for proteostasis maintenance.
Elife, 11, 2022
|
|
5JCL
 
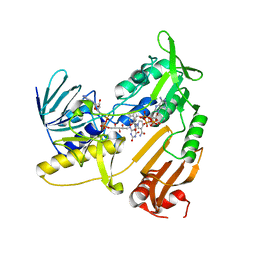 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Os09g0567300 protein | | Authors: | Park, A.K, Kim, H.W. | | Deposit date: | 2016-04-15 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
3OIX
 
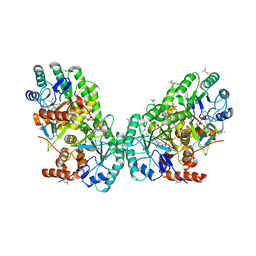 | | Crystal structure of the putative dihydroorotate dehydrogenase from Streptococcus mutans | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Putative dihydroorotate dehydrogenase; dihydroorotate oxidase | | Authors: | Liu, Y, Gao, Z.Q, Liu, C.P, Dong, Y.H. | | Deposit date: | 2010-08-20 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structure of the putative dihydroorotate dehydrogenase from Streptococcus mutans
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3OJW
 
 | | Disulfide crosslinked cytochrome P450 reductase inactive | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH-cytochrome P450 reductase | | Authors: | Xia, C, Hamdane, D, Shen, A, Choi, V, Kasper, C, Zhang, H, Im, S.-C, Waskell, L, Kim, J.-J.P. | | Deposit date: | 2010-08-23 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational Changes of NADPH-Cytochrome P450 Oxidoreductase Are Essential for Catalysis and Cofactor Binding.
J.Biol.Chem., 286, 2011
|
|
5JAP
 
 | |
5JIL
 
 | | Crystal structure of rat coronavirus strain New-Jersey Hemagglutinin-Esterase in complex with 4N-acetyl sialic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Bakkers, M.J.G, Feitsma, L.J, de Groot, R.J, Huizinga, E.G. | | Deposit date: | 2016-04-22 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4YNT
 
 | | Crystal structure of Aspergillus flavus FAD glucose dehydrogenase | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Glucose oxidase, putative | | Authors: | Yoshida, H, Sakai, G, Kojima, K, Kamitori, S, Sode, K. | | Deposit date: | 2015-03-11 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural analysis of fungus-derived FAD glucose dehydrogenase
Sci Rep, 5, 2015
|
|
5JLA
 
 | |
4N7Q
 
 | | Crystal structure of eukaryotic THIC from A. thaliana | | Descriptor: | COBALT (II) ION, HEXANE-1,6-DIOL, Phosphomethylpyrimidine synthase, ... | | Authors: | Coquille, S.C, Roux, C, Mehta, A, Begley, T.P, Fitzpatrick, T.B, Thore, S. | | Deposit date: | 2013-10-16 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution crystal structure of the eukaryotic HMP-P synthase (THIC) from Arabidopsis thaliana.
J.Struct.Biol., 184, 2013
|
|
4WHA
 
 | | Lipoxygenase-1 (soybean) L546A/L754A mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FE (III) ION, ... | | Authors: | Scouras, A.D, Carr, C.A.M, Hu, S, Klinman, J.P. | | Deposit date: | 2014-09-21 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extremely elevated room-temperature kinetic isotope effects quantify the critical role of barrier width in enzymatic C-H activation.
J.Am.Chem.Soc., 136, 2014
|
|
1RX0
 
 | | Crystal structure of isobutyryl-CoA dehydrogenase complexed with substrate/ligand. | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Acyl-CoA dehydrogenase family member 8, ... | | Authors: | Battaile, K.P, Nguyen, T.V, Vockley, J, Kim, J.J. | | Deposit date: | 2003-12-18 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures of Isobutyryl-CoA Dehydrogenase and Enzyme-Product Complex: COMPARISON WITH ISOVALERYL- AND SHORT-CHAIN ACYL-COA DEHYDROGENASES.
J.Biol.Chem., 279, 2004
|
|
3P68
 
 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | Descriptor: | GOLD 3+ ION, Lysozyme C | | Authors: | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
1S1F
 
 | | Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 from antibiotic biosynthetic pathways | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, GLYCEROL, MALONIC ACID, ... | | Authors: | Zhao, B, Lamb, D.C, Lei, L, Sundaramoorthy, M, Podust, L.M, Waterman, M.R. | | Deposit date: | 2004-01-06 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of Two Flaviolin Substrate Molecules, Oxidative Coupling, and Crystal Structure of Streptomyces coelicolor A3(2) Cytochrome P450 158A2
J.Biol.Chem., 280, 2005
|
|
3OQE
 
 | | Structure of OpdA mutant Y257F | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, Phosphotriesterase | | Authors: | Ely, F, Guddat, L.W, Ollis, D.L, Schenk, G. | | Deposit date: | 2010-09-02 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The organophosphate-degrading enzyme from Agrobacterium radiobacter displays mechanistic flexibility for catalysis.
Biochem.J., 432, 2010
|
|
1S20
 
 | | A novel NAD binding protein revealed by the crystal structure of E. Coli 2,3-diketogulonate reductase (YiaK) NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER82 | | Descriptor: | Hypothetical oxidoreductase yiaK, L(+)-TARTARIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Forouhar, F, Lee, I, Benach, J, Kulkarni, K, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-01-07 | | Release date: | 2004-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Novel NAD-binding Protein Revealed by the Crystal Structure of 2,3-Diketo-L-gulonate Reductase (YiaK).
J.Biol.Chem., 279, 2004
|
|
3P6S
 
 | |
4WG2
 
 | | P411BM3-CIS T438S I263F regioselective C-H amination catalyst | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Hyster, T.K, Farwell, C.C, Buller, A.R, McIntosh, J.A, Arnold, F.H. | | Deposit date: | 2014-09-17 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Enzyme-controlled nitrogen-atom transfer enables regiodivergent C-h amination.
J.Am.Chem.Soc., 136, 2014
|
|
1RY8
 
 | | Prostaglandin F synthase complexed with NADPH and rutin | | Descriptor: | Aldo-keto reductase family 1 member C3, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, RUTIN | | Authors: | Komoto, J, Yamada, T, Watanabe, K, Takusagawa, F. | | Deposit date: | 2003-12-19 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of human prostaglandin F synthase (AKR1C3).
Biochemistry, 43, 2004
|
|
4N1E
 
 | | Structural evidence for antigen receptor evolution | | Descriptor: | Lysozyme C, immunoglobulin variable light chain domain | | Authors: | Langley, D.B, Rouet, R, Stock, D, Christ, D. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-29 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural reconstruction of protein ancestry.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
