1DVL
 
 | |
1OME
 
 | | CRYSTAL STRUCTURE OF THE OMEGA LOOP DELETION MUTANT (RESIDUES 163-178 DELETED) OF BETA-LACTAMASE FROM STAPHYLOCOCCUS AUREUS PC1 | | Descriptor: | BETA-LACTAMASE, CHLORIDE ION | | Authors: | Banerjee, S, Pieper, U, Herzberg, O. | | Deposit date: | 1998-02-09 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of the omega-loop in the activity, substrate specificity, and structure of class A beta-lactamase.
Biochemistry, 37, 1998
|
|
1OMY
 
 | | Crystal Structure of a Recombinant alpha-insect Toxin BmKaIT1 from the scorpion Buthus martensii Karsch | | Descriptor: | ACETIC ACID, Alpha-neurotoxin TX12, CHLORIDE ION | | Authors: | Huang, Y, Huang, Q, Chen, H, Tang, Y, Miyake, H, Kusunoki, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization and preliminary crystallographic study of rBmKalphaIT1, a recombinant alpha-insect toxin from the scorpion Buthus martensii Karsch.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1J3C
 
 | |
1OR6
 
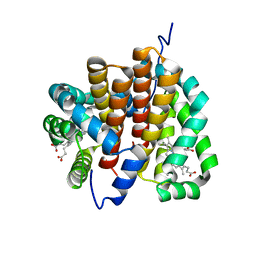 | |
1OTT
 
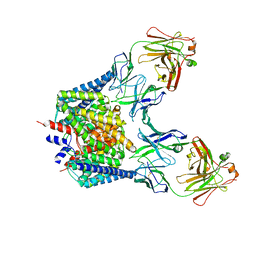 | | Structure of the Escherichia coli ClC Chloride channel E148A mutant and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (Heavy chain), Fab fragment (Light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-23 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
1OWI
 
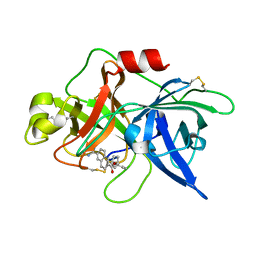 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-[3-(CYCLOPENTYLOXY)PHENYL]-2-NAPHTHAMIDE, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Rockway, T.W, Geyer, A, McClellan, W, Weitzberg, M, Zhao, X, Mantei, R, Nienaber, V.L, Stewart, K, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2003-03-28 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Identification of Novel Binding Interactions in the Development of Potent, Selective 2-Naphthamidine Inhibitors of Urokinase. Synthesis, Structural Analysis, and SAR of N-Phenyl Amide 6-Substitution.
J.Med.Chem., 47, 2004
|
|
1OQC
 
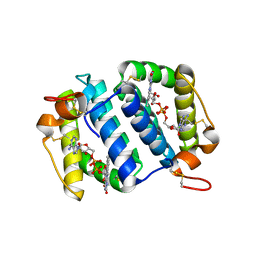 | |
1OYZ
 
 | | X-RAY STRUCTURE OF YIBA_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET31. | | Descriptor: | Hypothetical protein yibA | | Authors: | Kuzin, A, Semesi, A, Skarina, T, Savchenko, A, Arrowsmith, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-04-07 | | Release date: | 2003-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-RAY STRUCTURE OF YIBA_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET31.
To be Published
|
|
1E2A
 
 | | ENZYME IIA FROM THE LACTOSE SPECIFIC PTS FROM LACTOCOCCUS LACTIS | | Descriptor: | ENZYME IIA, MAGNESIUM ION | | Authors: | Sliz, P, Engelmann, R, Hengstenberg, W, Pai, E.F. | | Deposit date: | 1997-04-25 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of enzyme IIAlactose from Lactococcus lactis reveals a new fold and points to possible interactions of a multicomponent system.
Structure, 5, 1997
|
|
1P0E
 
 | | CRYSTAL STRUCTURE OF ZYMOMONAS MOBILIS tRNA-GUANINE TRANSGLYCOSYLASE (TGT) COCRYSTALLISED WITH PREQ1 AT PH 5.5 | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Stubbs, M.T, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2003-04-10 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Flexible adaptations in the structure of the tRNA-modifying enzyme tRNA-guanine transglycosylase
and their implications for substrate selectivity, reaction mechanism and structure-based drug design
Chembiochem, 4, 2003
|
|
1OSE
 
 | | Porcine pancreatic alpha-amylase complexed with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Gilles, C, Payan, F. | | Deposit date: | 1996-03-20 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of pig pancreatic alpha-amylase isoenzyme II, in complex with the carbohydrate inhibitor acarbose.
Eur.J.Biochem., 238, 1996
|
|
1P2U
 
 | | H-Ras in 50% isopropanol | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Buhrman, G.K, de Serrano, V, Mattos, C. | | Deposit date: | 2003-04-16 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Organic solvents order the dynamic switch II in Ras crystals
Structure, 11, 2003
|
|
1P4C
 
 | | High Resolution Structure of Oxidized Active Mutant of (S)-Mandelate Dehydrogenase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, L(+)-Mandelate Dehydrogenase, ... | | Authors: | Sukumar, N, Mitra, B, Mathews, F.S. | | Deposit date: | 2003-04-22 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High Resolution Structures of an Oxidized and Reduced Flavoprotein: THE WATER SWITCH IN A SOLUBLE FORM OF (S)-MANDELATE DEHYDROGENASE
J.Biol.Chem., 279, 2004
|
|
1P4U
 
 | | CRYSTAL STRUCTURE OF GGA3 GAE DOMAIN IN COMPLEX WITH RABAPTIN-5 PEPTIDE | | Descriptor: | ADP-ribosylation factor binding protein GGA3, Rabaptin-5 | | Authors: | Miller, G.J, Mattera, R, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2003-04-24 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RECOGNITION OF ACCESSORY PROTEIN MOTIFS BY THE GAMMA-ADAPTIN EAR DOMAIN OF GGA3
Nat.Struct.Biol., 10, 2003
|
|
1OWQ
 
 | | Crystal structure of a 40 kDa signalling protein (SPC-40) secreted during involution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, signal processing protein | | Authors: | Kumar, J, Sharma, S, Jasti, J, Bhushan, A, Singh, T.P. | | Deposit date: | 2003-03-29 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a 40 kDa signalling protein (SPC-40) secreted during involution
To be Published
|
|
1O3T
 
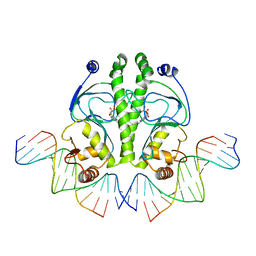 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | Descriptor: | 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*TP*CP*G)-3', 5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Chen, S, Vojtechovsky, J, Parkinson, G.N, Ebright, R.H, Berman, H.M. | | Deposit date: | 2003-03-18 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex:
DNA Binding Specificity Based on Energetics of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
1O4A
 
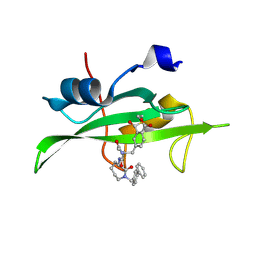 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU82197. | | Descriptor: | 4-[2-ACETYLAMINO-2-(1-BIPHENYL-4-YLMETHYL-2-OXO-AZEPAN-3-YLCARBAMOYL)-ETHYL]-2-FORMYL-BENZOIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1E4J
 
 | |
1NMA
 
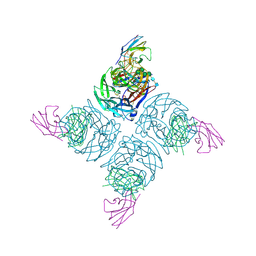 | | N9 NEURAMINIDASE COMPLEXES WITH ANTIBODIES NC41 AND NC10: EMPIRICAL FREE-ENERGY CALCULATIONS CAPTURE SPECIFICITY TRENDS OBSERVED WITH MUTANT BINDING DATA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB NC10, N9 NEURAMINIDASE, ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1994-05-06 | | Release date: | 1995-09-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | N9 neuraminidase complexes with antibodies NC41 and NC10: empirical free energy calculations capture specificity trends observed with mutant binding data.
Biochemistry, 33, 1994
|
|
1O9Q
 
 | |
1OAS
 
 | | O-ACETYLSERINE SULFHYDRYLASE FROM SALMONELLA TYPHIMURIUM | | Descriptor: | O-ACETYLSERINE SULFHYDRYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Burkhard, P, Rao, G.S.J, Hohenester, E, Schnackerz, K.D, Cook, P.F, Jansonius, J.N. | | Deposit date: | 1999-01-29 | | Release date: | 2000-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of O-acetylserine sulfhydrylase from Salmonella typhimurium.
J.Mol.Biol., 283, 1998
|
|
1NOB
 
 | | KNOB DOMAIN FROM ADENOVIRUS SEROTYPE 12 | | Descriptor: | PROTEIN (FIBER KNOB PROTEIN) | | Authors: | Bewley, M.C, Springer, K, Zhang, Y.B, Freimuth, P, Flanagan, J.M. | | Deposit date: | 1999-05-05 | | Release date: | 1999-11-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR.
Science, 286, 1999
|
|
1NP8
 
 | | 18-k C-terminally trunucated small subunit of calpain | | Descriptor: | CADMIUM ION, Calcium-dependent protease, small subunit | | Authors: | Leinala, E.K, Arthur, J.S, Grochulski, P, Davies, P.L, Elce, J.S, Jia, Z. | | Deposit date: | 2003-01-17 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A second binding site revealed by C-terminal truncation of calpain small subunit, a penta-EF-hand protein
PROTEINS: STRUCT.,FUNCT.,GENET., 53, 2003
|
|
1OCK
 
 | |
