1ZS2
 
 | | Amylosucrase Mutant E328Q in a ternary complex with sucrose and maltoheptaose | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, amylosucrase, ... | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, van der Veen, B.A, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2005-05-23 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of the Glu328Gln mutant of Neisseria polysaccharea amylosucrase in complex with sucrose and maltoheptaose
BIOCATAL.BIOTRANSFOR., 24, 2006
|
|
1TPS
 
 | |
1TYN
 
 | | ATOMIC STRUCTURE OF THE TRYPSIN-CYCLOTHEONAMIDE A COMPLEX: LESSONS FOR THE DESIGN OF SERINE PROTEASE INHIBITORS | | Descriptor: | BETA-TRYPSIN, CYCLOTHEONAMIDE A | | Authors: | Lee, A.Y, Hagihara, M, Karmacharya, R, Albers, M.W, Schreiber, S.L, Clardy, J. | | Deposit date: | 1994-09-19 | | Release date: | 1995-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic Structure of the Trypsin-Cyclotheonamide a Complex: Lessons for the Design of Serine Protease Inhibitors
J.Am.Chem.Soc., 115, 1993
|
|
1TNF
 
 | |
1ZRM
 
 | | CRYSTAL STRUCTURE OF THE REACTION INTERMEDIATE OF L-2-HALOACID DEHALOGENASE WITH 2-CHLORO-N-BUTYRATE | | Descriptor: | L-2-HALOACID DEHALOGENASE, butanoic acid | | Authors: | Li, Y.-F, Hata, Y, Fujii, T, Hisano, T, Nishihara, M, Kurihara, T, Esaki, N. | | Deposit date: | 1998-03-03 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of reaction intermediates of L-2-haloacid dehalogenase and implications for the reaction mechanism.
J.Biol.Chem., 273, 1998
|
|
2EK9
 
 | |
2E8I
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, D1 mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Shibata, N, Higuchi, Y, Negoro, S. | | Deposit date: | 2007-01-20 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold.
Febs J., 276, 2009
|
|
1UIH
 
 | | ANALYSIS OF THE STABILIZATION OF HEN LYSOZYME WITH THE HELIX DIPOLE AND CHARGED SIDE CHAINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Motoshima, H, Ohmura, T, Ueda, T, Imoto, T. | | Deposit date: | 1996-11-26 | | Release date: | 1997-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fluctuations in free or substrate-complexed lysozyme and a mutant of it detected on x-ray crystallography and comparison with those detected on NMR.
J.Biochem.(Tokyo), 131, 2002
|
|
2ACE
 
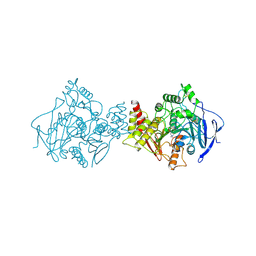 | | NATIVE ACETYLCHOLINESTERASE (E.C. 3.1.1.7) FROM TORPEDO CALIFORNICA | | Descriptor: | ACETYLCHOLINE, ACETYLCHOLINESTERASE | | Authors: | Harel, M, Raves, M.L, Silman, I, Sussman, J.L. | | Deposit date: | 1996-06-23 | | Release date: | 1996-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of acetylcholinesterase complexed with the nootropic alkaloid, (-)-huperzine A.
Nat.Struct.Biol., 4, 1997
|
|
1UIB
 
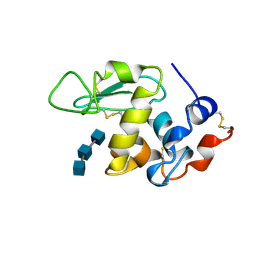 | | ANALYSIS OF THE STABILIZATION OF HEN LYSOZYME WITH THE HELIX DIPOLE AND CHARGED SIDE CHAINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Motoshima, H, Ohmura, T, Ueda, T, Imoto, T. | | Deposit date: | 1996-11-26 | | Release date: | 1997-11-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fluctuations in free or substrate-complexed lysozyme and a mutant of it detected on x-ray crystallography and comparison with those detected on NMR.
J.Biochem.(Tokyo), 131, 2002
|
|
2DYJ
 
 | | Crystal structure of ribosome-binding factor A from Thermus thermophilus HB8 | | Descriptor: | Ribosome-binding factor A | | Authors: | Kawazoe, M, Takemoto, C, Nakayama-Ushikoshi, R, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-14 | | Release date: | 2007-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural aspects of RbfA action during small ribosomal subunit assembly.
Mol.Cell, 28, 2007
|
|
2AD7
 
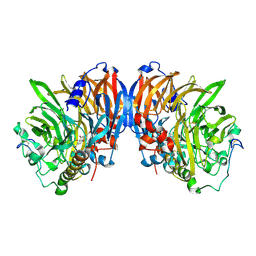 | | crystal structure of methanol dehydrogenase from M. W3A1 (form C) in the presence of methanol | | Descriptor: | CALCIUM ION, Methanol dehydrogenase subunit 1, Methanol dehydrogenase subunit 2, ... | | Authors: | Li, J, Gan, J.-H, Xia, Z.-X, Mathews, F.S. | | Deposit date: | 2005-07-20 | | Release date: | 2006-07-25 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The enzymatic reaction-induced configuration change of the prosthetic group PQQ of methanol dehydrogenase
Biochem.Biophys.Res.Commun., 406, 2011
|
|
1UHD
 
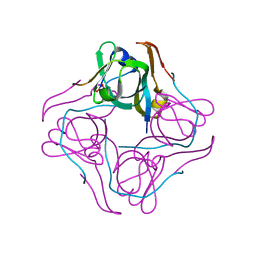 | | Crystal structure of aspartate decarboxylase, pyruvoly group bound form | | Descriptor: | Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain | | Authors: | Lee, B.I, Kwon, A.-R, Han, B.W, Suh, S.W. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the schiff base intermediate prior to decarboxylation in the catalytic cycle of aspartate alpha-decarboxylase
J.Mol.Biol., 340, 2004
|
|
2E00
 
 | |
1ZXE
 
 | | Crystal Structure of eIF2alpha Protein Kinase GCN2: D835N Inactivating Mutant in Apo Form | | Descriptor: | GLYCEROL, Serine/threonine-protein kinase | | Authors: | Padyana, A.K, Qiu, H, Roll-Mecak, A, Hinnebusch, A.G, Burley, S.K. | | Deposit date: | 2005-06-07 | | Release date: | 2005-06-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Autoinhibition and Mutational Activation of Eukaryotic Initiation Factor 2{alpha} Protein Kinase GCN2
J.Biol.Chem., 280, 2005
|
|
2ADU
 
 | | Human Methionine Aminopeptidase Complex with 4-Aryl-1,2,3-triazole Inhibitor | | Descriptor: | 4-(3-METHYLPHENYL)-1H-1,2,3-TRIAZOLE, COBALT (II) ION, Methionine aminopeptidase 2 | | Authors: | Kallander, L.S, Lu, Q, Chen, W, Tomaszek, T, Yang, G, Tew, D, Meek, T.D, Hofmann, G.A, Schulz-Pritchard, C.K, Smith, W.W, Janson, C.A, Ryan, M.D, Zhang, G.F, Johanson, K.O, Kirkpatrick, R.B, Ho, T.F, Fisher, P.W, Mattern, M.R, Johnson, R.K, Hansbury, M.J, Winkler, J.D, Ward, K.W, Veber, D.F, Thompson, S.K. | | Deposit date: | 2005-07-20 | | Release date: | 2005-09-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 4-Aryl-1,2,3-triazole: A Novel Template for a Reversible Methionine Aminopeptidase 2 Inhibitor, Optimized To Inhibit Angiogenesis in Vivo
J.Med.Chem., 48, 2005
|
|
2EIZ
 
 | | Crystal structure of humanized HYHEL-10 fv mutant(HW47Y)-hen lysozyme complex | | Descriptor: | ANTI-LYSOZYME ANTIBODY FV REGION, Lysozyme C | | Authors: | Nakanishi, T, Tsumoto, K, Yokota, A, Kondo, H, Kumagai, I. | | Deposit date: | 2007-03-14 | | Release date: | 2008-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Critical contribution of VH-VL interaction to reshaping of an antibody: the case of humanization of anti-lysozyme antibody, HyHEL-10
Protein Sci., 17, 2008
|
|
2E0I
 
 | | Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: Implication of a novel light-harvesting cofactor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 432aa long hypothetical deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fujihashi, M, Numoto, N, Kobayashi, Y, Mizushima, A, Tsujimura, M, Nakamura, A, Kawarabayashi, Y, Miki, K. | | Deposit date: | 2006-10-10 | | Release date: | 2006-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Archaeal Photolyase from Sulfolobus tokodaii with Two FAD Molecules: Implication of a Novel Light-harvesting Cofactor
J.Mol.Biol., 365, 2007
|
|
1UI1
 
 | | Crystal Structure Of Uracil-DNA Glycosylase From Thermus Thermophilus HB8 | | Descriptor: | IRON/SULFUR CLUSTER, Uracil-DNA Glycosylase | | Authors: | Hoseki, J, Okamoto, A, Masui, R, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-14 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Family 4 Uracil-DNA Glycosylase from Thermus thermophilus HB8
J.Mol.Biol., 333, 2003
|
|
1UII
 
 | |
1W6M
 
 | | X-RAY CRYSTAL STRUCTURE OF C2S HUMAN GALECTIN-1 COMPLEXED WITH GALACTOSE | | Descriptor: | BETA-MERCAPTOETHANOL, GALECTIN-1, SULFATE ION, ... | | Authors: | Lopez-Lucendo, M.I.F, Gabius, H.J, Romero, A. | | Deposit date: | 2004-08-19 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Growth-Regulatory Human Galectin-1: Crystallographic Characterisation of the Structural Changes Induced by Single-Site Mutations and Their Impact on the Thermodynamics of Ligand Binding.
J.Mol.Biol., 343, 2004
|
|
25C8
 
 | | CATALYTIC ANTIBODY 5C8, FAB-HAPTEN COMPLEX | | Descriptor: | IGG 5C8, N-METHYL-N-(PARA-GLUTARAMIDOPHENYL-ETHYL)-PIPERIDINIUM ION | | Authors: | Gruber, K, Wilson, I.A. | | Deposit date: | 1998-03-18 | | Release date: | 1999-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for antibody catalysis of a disfavored ring closure reaction.
Biochemistry, 38, 1999
|
|
1W8V
 
 | |
2EL9
 
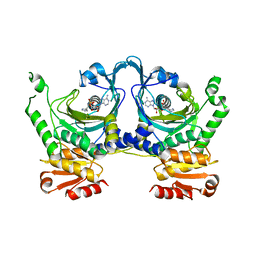 | | Crystal structure of E.coli Histidyl-tRNA synthetase complexed with a histidyl-adenylate analogue | | Descriptor: | 5'-O-[(L-HISTIDYLAMINO)SULFONYL]ADENOSINE, Histidyl-tRNA synthetase | | Authors: | Yanagisawa, T, Si, S.Y, Matsuno, M, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of E.coli Histidyl-tRNA synthetase complexed with a histidyl-adenylate analogue
To be published
|
|
1VTK
 
 | |
