5JLP
 
 | |
3KAA
 
 | | Structure of Tim-3 in complex with phosphatidylserine | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, CALCIUM ION, Hepatitis A virus cellular receptor 2 | | Authors: | Ballesteros, A, Santiago, C, Casasnovas, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | T cell/transmembrane, Ig, and mucin-3 allelic variants differentially recognize phosphatidylserine and mediate phagocytosis of apoptotic cells.
J.Immunol., 184, 2010
|
|
6WH9
 
 | | Ketoreductase from module 1 of the 6-deoxyerythronolide B synthase (KR1) in complex with antibody fragment (Fab) 1D10 | | Descriptor: | 1D10 (Fab heavy chain), 1D10 (Fab light chain), KR1, ... | | Authors: | Cogan, D.P, Mathews, I.I, Khosla, C. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Antibody Probes of Module 1 of the 6-Deoxyerythronolide B Synthase Reveal an Extended Conformation During Ketoreduction.
J.Am.Chem.Soc., 142, 2020
|
|
8GF2
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies eCR3022.20 and CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 Fab heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Broadening a SARS-CoV-1-neutralizing antibody for potent SARS-CoV-2 neutralization through directed evolution.
Sci.Signal., 16, 2023
|
|
8JK8
 
 | | SP1746 in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, GLYCEROL, ... | | Authors: | Jin, Y, Niu, L, Ke, J. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia.
Structure, 32, 2024
|
|
8JKR
 
 | | SP1746 in complex with UMP | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, GLYCEROL, ... | | Authors: | Jin, Y, Niu, L, Ke, J. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia.
Structure, 32, 2024
|
|
4U5P
 
 | | Crystal structure of native RhCC (YP_702633.1) from Rhodococcus jostii RHA1 at 1.78 Angstrom | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Poddar, H, Rozeboom, H.J, Thunnissen, A.M.W.H. | | Deposit date: | 2014-07-25 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Functional and structural characterization of an unusual cofactor-independent oxygenase.
Biochemistry, 54, 2015
|
|
7NU9
 
 | | Crystal Structure of Neisseria gonorrhoeae LeuRS in Complex with ATP and L-leucinol | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-4-METHYL-PENTAN-1-OL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2021-03-11 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Partitioning of the initial catalytic steps of leucyl-tRNA synthetase is driven by an active site peptide-plane flip.
Commun Biol, 5, 2022
|
|
7YT0
 
 | |
7NUC
 
 | | Crystal Structure of Neisseria gonorrhoeae LeuRS L550G mutant in Complex with ATP and L-leucinol | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-4-METHYL-PENTAN-1-OL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2021-03-11 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Partitioning of the initial catalytic steps of leucyl-tRNA synthetase is driven by an active site peptide-plane flip.
Commun Biol, 5, 2022
|
|
7NU3
 
 | | Crystal Structure of Neisseria gonorrhoeae LeuRS E169G mutant in Complex with ATP and L-leucinol | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-4-METHYL-PENTAN-1-OL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2021-03-11 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Partitioning of the initial catalytic steps of leucyl-tRNA synthetase is driven by an active site peptide-plane flip.
Commun Biol, 5, 2022
|
|
7N8W
 
 | |
8Q1C
 
 | | Substrate-free D10N,P146A variant of beta-phosphoglucomutase from Lactococcus lactis | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-phosphoglucomutase, ... | | Authors: | Cruz-Navarrete, F.A, Baxter, N.J, Flinders, A.J, Buzoianu, A, Cliff, M.J, Baker, P.J, Waltho, J.P. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Peri active site catalysis of proline isomerisation is the molecular basis of allomorphy in beta-phosphoglucomutase.
Commun Biol, 7, 2024
|
|
7N93
 
 | | P70 S6K1 IN COMPLEX WITH MSC2363318A-1 | | Descriptor: | 4-({(1S)-2-(azetidin-1-yl)-1-[4-chloro-3-(trifluoromethyl)phenyl]ethyl}amino)quinazoline-8-carboxamide, Ribosomal protein S6 kinase beta-1 | | Authors: | Mochalkin, I. | | Deposit date: | 2021-06-16 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Identification of Clinical Candidate M2698, a Dual p70S6K and Akt Inhibitor, for Treatment of PAM Pathway-Altered Cancers.
J.Med.Chem., 64, 2021
|
|
8UHH
 
 | | anti-Phosphohistidine Fab hSC44.ck.elbow.20.N32F | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Hunter, T, Wilson, I.A. | | Deposit date: | 2023-10-09 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody.
Biorxiv, 2024
|
|
8Q23
 
 | | HsNMT1 in complex with both MyrCoA and Ac-D-ORN-SFSKPR inhibitor peptide | | Descriptor: | ACE-ORD-SER-PHE-SER-LYS-PRO-ARG, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-08-14 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel, tightly structurally related N-myristoyltransferase inhibitors display equally potent yet distinct inhibitory mechanisms.
Structure, 32, 2024
|
|
6YQQ
 
 | | ForT-PRPP complex | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, CHLORIDE ION, ForT-PRPP complex, ... | | Authors: | Naismith, J.H, Gao, S. | | Deposit date: | 2020-04-18 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Uncovering the chemistry of C-C bond formation in C-nucleoside biosynthesis: crystal structure of a C-glycoside synthase/PRPP complex.
Chem.Commun.(Camb.), 56, 2020
|
|
8GO3
 
 | |
8Q24
 
 | | HsNMT1 in complex with both MyrCoA and (DAB)SFSKPR inhibitor peptide | | Descriptor: | CHLORIDE ION, DAB-SER-PHE-SER-LYS-PRO-ARG, GLYCEROL, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-08-14 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel, tightly structurally related N-myristoyltransferase inhibitors display equally potent yet distinct inhibitory mechanisms.
Structure, 32, 2024
|
|
9DX7
 
 | | LRRC8A:D Conformation 1 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8D | | Authors: | Lurie, A, Brohawn, S.G. | | Deposit date: | 2024-10-10 | | Release date: | 2024-11-27 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of a LRRC8A:D volume-regulated anion channel.
To Be Published
|
|
7NM0
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1-(2,6-dihydroxyphenyl)ethan-1-one. | | Descriptor: | 1,2-ETHANEDIOL, 1-[2,6-bis(oxidanyl)phenyl]ethanone, Acetylglutamate kinase, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
6YOA
 
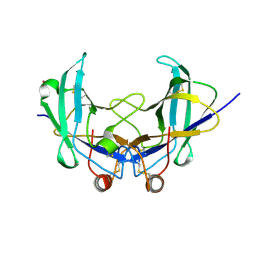 | | Lig v 1 structure and the inflammatory response to the Ole e 1 protein family | | Descriptor: | Major pollen allergen Lig v 1, NICKEL (II) ION | | Authors: | Robledo-Retana, T, Bradley-Clark, J, Croll, T, Rose, R, Stagg, A, Villalba, M, Pickersgill, R. | | Deposit date: | 2020-04-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Lig v 1 structure and the inflammatory response to the Ole e 1 protein family.
Allergy, 75, 2020
|
|
4Y24
 
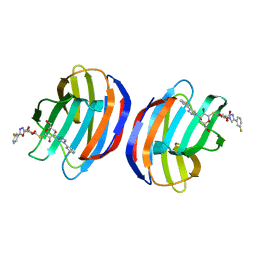 | | Complex of human Galectin-1 and TD-139 | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, Galectin-1 | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors.
Sci Rep, 6, 2016
|
|
7NLU
 
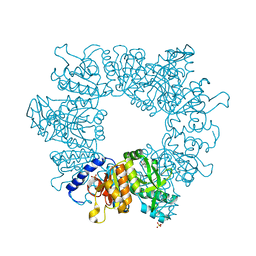 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1-(1H-indol-3-yl)ethan-1-one | | Descriptor: | 1-(1~{H}-indol-3-yl)ethanone, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.235 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7QNY
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with COVOX-58 and COVOX-158 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 heavy chain, COVOX-158 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
