7MFF
 
 | | Dimeric (BRAF)2:(14-3-3)2 complex bound to SB590885 Inhibitor | | Descriptor: | (1Z)-5-(2-{4-[2-(DIMETHYLAMINO)ETHOXY]PHENYL}-5-PYRIDIN-4-YL-1H-IMIDAZOL-4-YL)INDAN-1-ONE OXIME, 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Martinez Fiesco, J.A, Ping, Z, Durrant, D.E, Morrison, D.K. | | Deposit date: | 2021-04-09 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Structural insights into the BRAF monomer-to-dimer transition mediated by RAS binding.
Nat Commun, 13, 2022
|
|
4RVL
 
 | | CHK1 kinase domain with diazacarbazole compound 7: 3-(2-hydroxyphenyl)-9H-pyrrolo[2,3-b:5,4-c']dipyridine-6-carbonitrile | | Descriptor: | 3-(2-hydroxyphenyl)-9H-pyrrolo[2,3-b:5,4-c']dipyridine-6-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Wiesmann, C, Wu, P. | | Deposit date: | 2014-11-26 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mitigation of Acetylcholine Esterase Activity in the 1,7-Diazacarbazole Series of Inhibitors of Checkpoint Kinase 1.
J.Med.Chem., 58, 2015
|
|
4XHD
 
 | | STRUCTURE OF HUMAN PREGNANE X RECEPTOR LIGAND BINDING DOMAIN WITH COMPOUND-1 | | Descriptor: | GLYCEROL, N-{(2R)-1-[(4S)-4-(4-chlorophenyl)-4-hydroxy-3,3-dimethylpiperidin-1-yl]-3-methyl-1-oxobutan-2-yl}-2-cyclopropylacetamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Khan, J.A, Camac, D.M. | | Deposit date: | 2015-01-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Developing Adnectins That Target SRC Co-Activator Binding to PXR: A Structural Approach toward Understanding Promiscuity of PXR.
J.Mol.Biol., 427, 2015
|
|
5JLR
 
 | | Crystal structure of Mycobacterium avium SerB2 with serine present at slightly different position near ACT domain | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, MAGNESIUM ION, ... | | Authors: | Shree, S, Agrawal, A, Dubey, S, Ramachandran, R. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Crystal structure of Mycobacterium avium SerB2 with serine present at slightly different position near ACT domain
To be published
|
|
8P1S
 
 | | Bifidobacterium asteroides alpha-L-fucosidase (TT1819) in complex with fucose. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifidobacterium asteroides alpha-L-fucosidase (TT1819), ... | | Authors: | Owen, C.D, Penner, M, Gascuena, A.N, Wu, H, Hernando, P.J, Monaco, S, Le Gall, G, Gardner, R, Ndeh, D, Urbanowicz, P.A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring the sequence-function space of microbial fucosidases.
Commun Chem, 7, 2024
|
|
9F3G
 
 | | Human carbonic anhydrase XII with 3-(cyclooctylamino)-2,6-difluoro-5-((4-hydroxybutyl)amino)-4-((3-hydroxypropyl)sulfonyl)benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 3-(cyclooctylamino)-2,6-bis(fluoranyl)-5-(4-oxidanylbutylamino)-4-[(~{E})-3-oxidanylprop-1-enyl]sulfonyl-benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Manakova, E, Grazulis, S, Smirnov, A, Paketuryte, V. | | Deposit date: | 2024-04-25 | | Release date: | 2025-05-14 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Di- meta -Substituted Fluorinated Benzenesulfonamides as Potent and Selective Anticancer Inhibitors of Carbonic Anhydrase IX and XII.
J.Med.Chem., 68, 2025
|
|
8P5B
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 500 micromolar X77 enantiomer S. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
6XHN
 
 | | Covalent complex of SARS-CoV main protease with 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-3-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | Descriptor: | (3S)-3-{[N-(4-methoxy-1H-indole-2-carbonyl)-L-leucyl]amino}-2-oxo-4-[(3S)-2-oxopyrrolidin-3-yl]butyl 2-cyanobenzoate, 1,2-ETHANEDIOL, 3C-like proteinase | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
4RT0
 
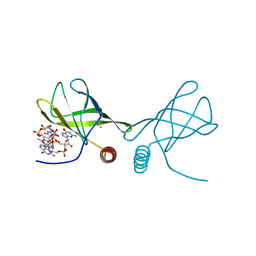 | | Structure of the Alg44 PilZ domain from Pseudomonas aeruginosa PAO1 in complex with c-di-GMP | | Descriptor: | 1,2-ETHANEDIOL, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Alginate biosynthesis protein Alg44 | | Authors: | Whitfield, G.B, Whitney, J.C. | | Deposit date: | 2014-11-12 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dimeric c-di-GMP Is Required for Post-translational Regulation of Alginate Production in Pseudomonas aeruginosa.
J.Biol.Chem., 290, 2015
|
|
1AXU
 
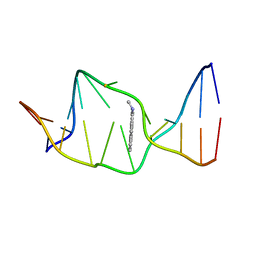 | | SOLUTION NMR STRUCTURE OF THE [AP]DG ADDUCT OPPOSITE DA IN A DNA DUPLEX, NMR, 9 STRUCTURES | | Descriptor: | DNA DUPLEX D(CCATC-[AP]G-CTACC)D(GGTAGAGATGG), N-1-AMINOPYRENE | | Authors: | Gu, Z, Gorin, A.A, Krishnasami, R, Hingerty, B.E, Basu, A.K, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dA in a DNA duplex.
Biochemistry, 38, 1999
|
|
8TJJ
 
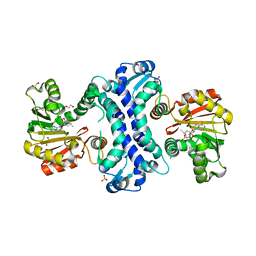 | | SAM-dependent methyltransferase RedM bound to SAM | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, RedM, ... | | Authors: | Daniel-Ivad, P, Ryan, K.S. | | Deposit date: | 2023-07-22 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of methyltransferase RedM that forms the dimethylpyrrolinium of the bisindole reductasporine.
J.Biol.Chem., 300, 2023
|
|
8GB1
 
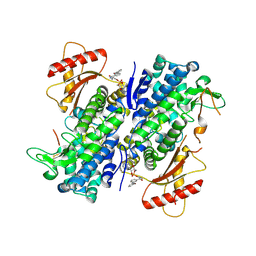 | | Crystal structure of SAMHD1 dimer bound to deoxyguanosine linked inhibitor | | Descriptor: | 5'-O-[(R)-(3-{[(1M)-3'-bromo[1,1'-biphenyl]-3-carbonyl]amino}propoxy)(hydroxy)phosphoryl]-2'-deoxyguanosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION | | Authors: | Egleston, M, Dong, L, Howlader, A.H, Bhat, S, Orris, B, Bianchet, M.A, Greenberg, M.M, Stivers, J.T. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Deoxyguanosine-Linked Bifunctional Inhibitor of SAMHD1 dNTPase Activity and Nucleic Acid Binding.
Acs Chem.Biol., 18, 2023
|
|
6WE8
 
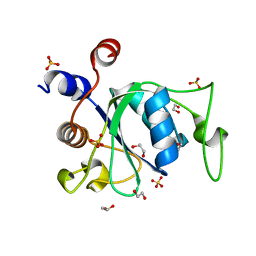 | | YTH domain of human YTHDC1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-01 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Biochemical and structural basis for YTH domain of human YTHDC1 binding to methylated adenine in DNA.
Nucleic Acids Res., 48, 2020
|
|
1EAZ
 
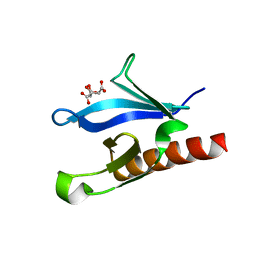 | | Crystal structure of the phosphoinositol (3,4)-bisphosphate binding PH domain of TAPP1 from human. | | Descriptor: | CITRIC ACID, TANDEM PH DOMAIN CONTAINING PROTEIN-1 | | Authors: | Thomas, C.C, Dowler, S, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2001-07-17 | | Release date: | 2002-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the Phosphatidylinositol 3,4-Bisphosphate-Binding Pleckstrin Homology (Ph) Domain of Tandem Ph-Domain-Containing Protein 1 (Tapp1): Molecular Basis of Lipid Specificity
Biochem.J., 358, 2001
|
|
6Z65
 
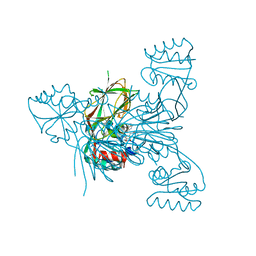 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | CITRIC ACID, NAD kinase 1, ~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-4-azanyl-butanamide | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2020-05-27 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | New Chemical Probe Targeting Bacterial NAD Kinase.
Molecules, 25, 2020
|
|
8VEY
 
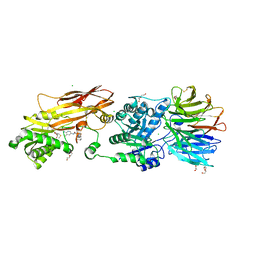 | | Crystal structure of PRMT5:MEP50 in complex with MTA and TNG908 | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
8VEU
 
 | |
5O00
 
 | |
8VHD
 
 | | Crystal Structure of Human IDH1 R132Q in complex with NADPH and Isocitrate | | Descriptor: | CALCIUM ION, GLYCEROL, IODIDE ION, ... | | Authors: | Mealka, M, Sohl, C.D, Huxford, T. | | Deposit date: | 2023-12-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
6FH3
 
 | | Protein arginine kinase McsB in the pArg-bound state | | Descriptor: | 1,2-ETHANEDIOL, Protein-arginine kinase, phospho-arginine | | Authors: | Suskiewicz, M.J, Heuck, A, Vu, L.D, Clausen, T. | | Deposit date: | 2018-01-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of McsB, a protein kinase for regulated arginine phosphorylation.
Nat.Chem.Biol., 15, 2019
|
|
6X9X
 
 | |
8BS7
 
 | |
4Y1X
 
 | | Complex of human Galectin-1 and Galbeta1-4(6OSO3)GlcNAc | | Descriptor: | Galectin-1, beta-D-galactopyranose-(1-4)-methyl 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranoside | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
6HLY
 
 | | Structure in P212121 form of the PBP AgtB in complex with agropinic acid from A.tumefacien R10 | | Descriptor: | 1,2-ETHANEDIOL, Agropine permease, agropinic acid | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
8E06
 
 | | Symmetry expansion of dimeric LRRK1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 1 | | Authors: | Reimer, J.M, Lin, Y.X, Leschziner, A.E. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of LRRK1 and mechanisms of autoinhibition and activation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
