1G4M
 
 | | CRYSTAL STRUCTURE OF BOVINE BETA-ARRESTIN 1 | | Descriptor: | BETA-ARRESTIN1 | | Authors: | Schubert, C, Han, M. | | Deposit date: | 2000-10-27 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane Translocation.
Structure, 9, 2001
|
|
1JSY
 
 | | Crystal structure of bovine arrestin-2 | | Descriptor: | Bovine arrestin-2 (full length) | | Authors: | Milano, S.K, Pace, H.C, Kim, Y.M, Brenner, C, Benovic, J.L. | | Deposit date: | 2001-08-19 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis.
Biochemistry, 41, 2002
|
|
6IF2
 
 | | Complex structure of Rab35 and its effector RUSC2 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Iporin, MAGNESIUM ION, ... | | Authors: | Lin, L, Zhu, J, Zhang, R. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rab35/ACAP2 and Rab35/RUSC2 Complex Structures Reveal Molecular Basis for Effector Recognition by Rab35 GTPase.
Structure, 27, 2019
|
|
6IF3
 
 | | Complex structure of Rab35 and its effector ACAP2 | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lin, L, Zhu, J, Zhang, R. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rab35/ACAP2 and Rab35/RUSC2 Complex Structures Reveal Molecular Basis for Effector Recognition by Rab35 GTPase.
Structure, 27, 2019
|
|
6U1N
 
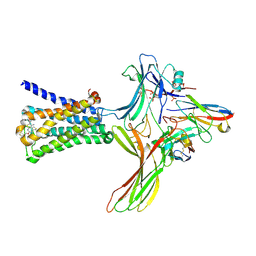 | | GPCR-Beta arrestin structure in lipid bilayer | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, Beta-arrestin-1, Fab30 heavy chain, ... | | Authors: | Staus, D.P, Hu, H, Robertson, M.J, Kleinhenz, A.L.W, Wingler, L.M, Capel, W.D, Latorraca, N.R, Lefkowitz, R.J, Skiniotis, G. | | Deposit date: | 2019-08-16 | | Release date: | 2020-02-26 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the M2 muscarinic receptor-beta-arrestin complex in a lipid nanodisc.
Nature, 579, 2020
|
|
2WTR
 
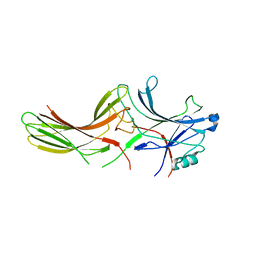 | |
8J8Z
 
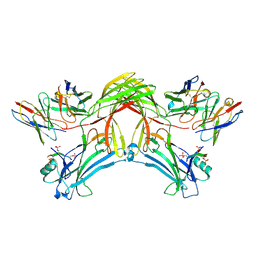 | | Structure of beta-arrestin1 in complex with D6Rpp | | Descriptor: | Atypical chemokine receptor 2, Beta-arrestin-1, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
3L81
 
 | | Crystal structure of adaptor protein complex 4 (AP-4) mu4 subunit C-terminal domain, in complex with a sorting peptide from the amyloid precursor protein (APP) | | Descriptor: | AP-4 complex subunit mu-1, Amyloid beta A4 protein, GLYCEROL | | Authors: | Mardones, G.A, Rojas, A.L, Burgos, P.V, Dasilva, L.L.P, Prabhu, Y, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2009-12-29 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sorting of the Alzheimer's disease amyloid precursor protein mediated by the AP-4 complex.
Dev.Cell, 18, 2010
|
|
6KL7
 
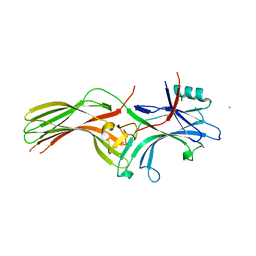 | | Beta-arrestin 1 mutant S13D/T275D | | Descriptor: | 1,2-ETHANEDIOL, BARIUM ION, Beta-arrestin-1 | | Authors: | Kang, H, Choi, H.J. | | Deposit date: | 2019-07-29 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Conformational Dynamics and Functional Implications of Phosphorylated beta-Arrestins.
Structure, 28, 2020
|
|
3LGE
 
 | | Crystal structure of rabbit muscle aldolase-SNX9 LC4 complex | | Descriptor: | Fructose-bisphosphate aldolase A, Sorting nexin-9 | | Authors: | Rangarajan, E.S, Park, H, Fortin, E, Sygusch, J, Izard, T. | | Deposit date: | 2010-01-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of aldolase control of sorting nexin 9 function in endocytosis.
J.Biol.Chem., 285, 2010
|
|
3MNM
 
 | | Crystal structure of GAE domain of GGA2p from Saccharomyces cerevisiae | | Descriptor: | ADP-ribosylation factor-binding protein GGA2, GLYCEROL, SULFATE ION | | Authors: | Fang, P, Wang, J, Li, X, Niu, L, Teng, M. | | Deposit date: | 2010-04-21 | | Release date: | 2010-09-08 | | Last modified: | 2014-03-05 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural basis for the specificity of the GAE domain of yGGA2 for its accessory proteins Ent3 and Ent5
Biochemistry, 49, 2010
|
|
1BXX
 
 | |
4JQI
 
 | | Structure of active beta-arrestin1 bound to a G protein-coupled receptor phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-arrestin-1, CHLORIDE ION, ... | | Authors: | Shukla, A.K, Manglik, A, Kruse, A.C, Xiao, K, Reis, R.I, Tseng, W.C, Staus, D.P, Hilger, D, Uysal, S, Huang, L.H, Paduch, M, Shukla, P.T, Koide, A, Koide, S, Weis, W.I, Kossiakoff, A.A, Kobilka, B.K, Lefkowitz, R.J. | | Deposit date: | 2013-03-20 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide.
Nature, 497, 2013
|
|
1BW8
 
 | |
1EDU
 
 | | CRYSTAL STRUCTURE OF THE ENTH DOMAIN OF RAT EPSIN 1 | | Descriptor: | 1,2-ETHANEDIOL, EH domain binding protein EPSIN | | Authors: | Hyman, J.H, Chen, H, Decamilli, P, Brunger, A.T. | | Deposit date: | 2000-01-28 | | Release date: | 2000-05-10 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Epsin 1 undergoes nucleocytosolic shuttling and its eps15 interactor NH(2)-terminal homology (ENTH) domain, structurally similar to Armadillo and HEAT repeats, interacts with the transcription factor promyelocytic leukemia Zn(2)+ finger protein (PLZF).
J.Cell Biol., 149, 2000
|
|
5X5P
 
 | | Human serum transferrin bound to ruthenium NTA | | Descriptor: | FE (III) ION, MALONATE ION, NITRILOTRIACETIC ACID, ... | | Authors: | Sun, H, Wang, M. | | Deposit date: | 2017-02-17 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding of ruthenium and osmium at non‐iron sites of transferrin accounts for their iron-independent cellular uptake.
J.Inorg.Biochem., 234, 2022
|
|
5Y6K
 
 | | Human serum trnasferrin bound to a fluorescent probe | | Descriptor: | (2S)-6-[2-(7-azido-4-methyl-2-oxidanylidene-chromen-3-yl)ethanoylamino]-2-[bis(2-hydroxy-2-oxoethyl)amino]hexanoic acid, FE (III) ION, MALONATE ION, ... | | Authors: | Jiang, N, Cheng, T, Wang, M, Chan, G.C.F, Jin, L, Li, H, Sun, H. | | Deposit date: | 2017-08-12 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Tracking iron-associated proteomes in pathogens by a fluorescence approach.
Metallomics, 10, 2018
|
|
5YQG
 
 | | The structure of 14-3-3 and pNumb peptide | | Descriptor: | 14-3-3 protein eta, Peptide from Protein numb homolog | | Authors: | Chen, X, Liu, Z, Wen, W. | | Deposit date: | 2017-11-06 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural determinants controlling 14-3-3 recruitment to the endocytic adaptor Numb and dissociation of the Numb/alpha-adaptin complex.
J. Biol. Chem., 293, 2018
|
|
1PZD
 
 | | Structural Identification of a conserved appendage domain in the carboxyl-terminus of the COPI gamma-subunit. | | Descriptor: | Coatomer gamma subunit, SULFATE ION | | Authors: | Hoffman, G.R, Rahl, P.B, Collins, R.N, Cerione, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Conserved Structural Motifs in Intracellular Trafficking Pathways. Structure of the gammaCOP Appendage Domain.
Mol.Cell, 12, 2003
|
|
8GO8
 
 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C5a anaphylatoxin chemotactic receptor 1, C5aR1 | | Descriptor: | Beta-arrestin-1, C5a anaphylatoxin chemotactic receptor 1, Fab30 heavy chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure of beta-arrestin in complex with a phosphopeptide
Mol.Cell, 2023
|
|
8GP3
 
 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C-X-C chemokine receptor type 4, CXCR4 | | Descriptor: | Beta-arrestin-1, C-X-C chemokine receptor type 4, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of beta-arrestin in complex with a phosphopeptide
Mol.Cell, 2023
|
|
8HSV
 
 | | The structure of rat beta-arrestin1 in complex with a rat Mdm2 peptide | | Descriptor: | Beta-arrestin-1, SULFATE ION, peptide from E3 ubiquitin-protein ligase Mdm2 | | Authors: | Yun, Y, Yoon, H.J, Choi, Y, Lee, H.H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GPCR targeting of E3 ubiquitin ligase MDM2 by inactive beta-arrestin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HST
 
 | |
6EHO
 
 | | Dimer of the Sortilin Vps10p domain at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thirup, S.S, Quistgaard, E.H, Januliene, D, Andersen, J.L, Nielsen, J.A. | | Deposit date: | 2017-09-14 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Acidic Environment Induces Dimerization and Ligand Binding Site Collapse in the Vps10p Domain of Sortilin.
Structure, 25, 2017
|
|
6ENR
 
 | |
