7ER0
 
 | |
7EQW
 
 | |
8C49
 
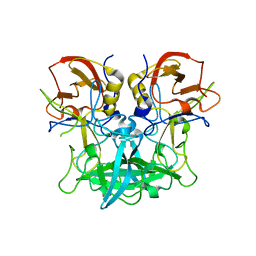
 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for 3-Methyl-L-histidine, bound to AMPPNP | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Hardy, F.J, Levy, C.W. | | Deposit date: | 2023-01-03 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Engineering mutually orthogonal PylRS/tRNA pairs for dual encoding of functional histidine analogues.
Protein Sci., 32, 2023
|
|
1RD7
 
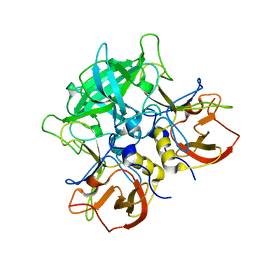
 | | DIHYDROFOLATE REDUCTASE COMPLEXED WITH FOLATE | | Descriptor: | BETA-MERCAPTOETHANOL, DIHYDROFOLATE REDUCTASE, FOLIC ACID | | Authors: | Sawaya, M.R, Kraut, J. | | Deposit date: | 1996-11-01 | | Release date: | 1996-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence.
Biochemistry, 36, 1997
|
|
2JDS
 
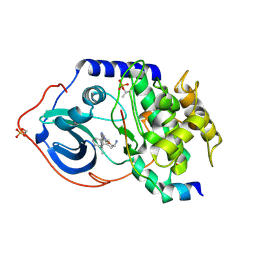 | | Structure of cAMP-dependent protein kinase complexed with A-443654 | | Descriptor: | (2S)-1-(1H-INDOL-3-YL)-3-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}PROPAN-2-AMINE, CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA | | Authors: | Davies, T.G, Verdonk, M.L, Graham, B, Saalau-Bethell, S, Hamlett, C.C.F, McHardy, T, Collins, I, Garrett, M.D, Workman, P, Woodhead, S.J, Jhoti, H, Barford, D. | | Deposit date: | 2007-01-12 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Comparison of Inhibitor Binding to Pkb, Pka and Pka-Pkb Chimera
J.Mol.Biol., 367, 2007
|
|
5HI5
 
 | | Binding site elucidation and structure guided design of macrocyclic IL-17A antagonists | | Descriptor: | (4S,20R)-7-chloro-N-methyl-4-{[(1-methyl-1H-pyrazol-5-yl)carbonyl]amino}-3,18-dioxo-2,19-diazatetracyclo[20.2.2.1~6,10~.1~11,15~]octacosa-1(24),6(28),7,9,11(27),12,14,22,25-nonaene-20-carboxamide, CAT-2000 FAB heavy chain, CAT-2000 light chain, ... | | Authors: | Liu, S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site elucidation and structure guided design of macrocyclic IL-17A antagonists.
Sci Rep, 6, 2016
|
|
1OZQ
 
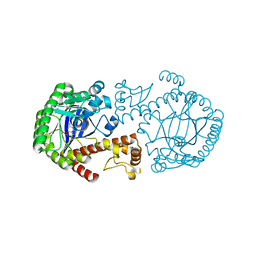 | | CRYSTAL STRUCTURE OF THE MUTATED TRNA-GUANINE TRANSGLYCOSYLASE (TGT)Y106F COMPLEXED WITH PREQ1 | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Stubbs, M.T, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2003-04-09 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flexible adaptations in the structure of the tRNA-modifying enzyme tRNA-guanine transglycosylase
and their implications for substrate selectivity, reaction mechanism and structure-based drug design
Chembiochem, 4, 2003
|
|
3DZ5
 
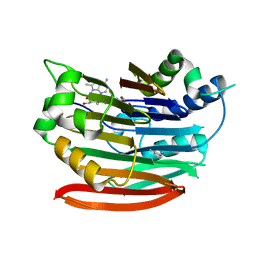 | | Human AdoMetDC with covalently bound 5'-[(2-aminooxyethyl)methylamino]-5'-deoxy-8-methyladenosine | | Descriptor: | 1,4-DIAMINOBUTANE, 5'-{[2-(aminooxy)ethyl](methyl)amino}-5'-deoxy-8-methyladenosine, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Bale, S, McCloskey, D.E, Pegg, A.E, Secrist III, J.A, Guida, W.C, Ealick, S.E. | | Deposit date: | 2008-07-29 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | New Insights into the Design of Inhibitors of Human S-Adenosylmethionine Decarboxylase: Studies of Adenine C8 Substitution in Structural Analogues of S-Adenosylmethionine
J.Med.Chem., 52, 2009
|
|
8GDI
 
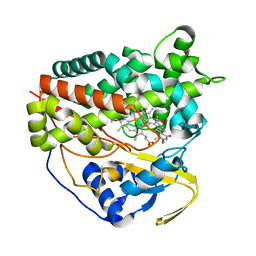 | | X-ray crystal structure of CYP124A1 from Mycobacterium Marinum in complex with 7-ketocholesterol | | Descriptor: | (3beta,8alpha,9beta)-3-hydroxycholest-5-en-7-one, Cytochrome P450 124A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ghith, A, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-03-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The oxidation of cholesterol derivatives by the CYP124 and CYP142 enzymes from Mycobacterium marinum.
J.Steroid Biochem.Mol.Biol., 231, 2023
|
|
2JDT
 
 | | Structure of PKA-PKB chimera complexed with ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY) ETHYLAMINO)ETHYL)AMIDE | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY)ETHYLAMINO)ETHYL)AMIDE | | Authors: | Davies, T.G, Verdonk, M.L, Graham, B, Saalau-Bethell, S, Hamlett, C.C.F, McHardy, T, Collins, I, Garrett, M.D, Workman, P, Woodhead, S.J, Jhoti, H, Barford, D. | | Deposit date: | 2007-01-12 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Structural Comparison of Inhibitor Binding to Pkb, Pka and Pka-Pkb Chimera
J.Mol.Biol., 367, 2007
|
|
1FPY
 
 | |
1OZM
 
 | | Y106F mutant of Z. mobilis TGT | | Descriptor: | Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Stubbs, M.T, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2003-04-09 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Flexible adaptations in the structure of the tRNA-modifying enzyme
tRNA-guanine transglycosylase and their implications for substrate selectivity,
reaction mechanism and structure-based drug design
Chembiochem, 4, 2003
|
|
1P0B
 
 | | Crystal Structure Of tRNA-Guanine Transglycosylase (TGT) From Zymomonas mobilis Complexed With Archaeosine Precursor, Preq0 | | Descriptor: | 2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDINE-5-CARBONITRILE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Stubbs, M.T, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2003-04-10 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Flexible adaptations in the structure of the tRNA-modifying enzyme tRNA-guanine transglycosylase
and their implications for substrate selectivity, reaction mechanism and structure-based drug design
Chembiochem, 4, 2003
|
|
5KXI
 
 | | X-ray structure of the human Alpha4Beta2 nicotinic receptor | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuronal acetylcholine receptor subunit alpha-4, ... | | Authors: | Morales-Perez, C.L, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.941 Å) | | Cite: | X-ray structure of the human alpha 4 beta 2 nicotinic receptor.
Nature, 538, 2016
|
|
3DL5
 
 | | Crystal Structure of the A287F Active Site Mutant of TS-DHFR from Cryptosporidium hominis | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DIHYDROFOLIC ACID, ... | | Authors: | Vargo, M.A, Martucci, W.E, Anderson, K.S. | | Deposit date: | 2008-06-26 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Explaining an unusually fast parasitic enzyme: folate tail-binding residues dictate substrate positioning and catalysis in Cryptosporidium hominis thymidylate synthase.
Biochemistry, 47, 2008
|
|
6H29
 
 | | HUMAN CARBONIC ANHYDRASE II IN COMPLEX WITH BENZYL CARBAMATE | | Descriptor: | (phenylmethyl) carbamate, Carbonic anhydrase 2, ZINC ION | | Authors: | Alterio, V, De Simone, G. | | Deposit date: | 2018-07-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Inhibition of carbonic anhydrases by a substrate analog: benzyl carbamate directly coordinates the catalytic zinc ion mimicking bicarbonate binding.
Chem. Commun. (Camb.), 54, 2018
|
|
3ARK
 
 | | Cl- binding hemoglobin component V form Propsilocerus akamusi under 1 M NaCl at pH 4.6 | | Descriptor: | CHLORIDE ION, Hemoglobin V, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kuwada, T, Hasegawa, T, Takagi, T, Shishikura, F. | | Deposit date: | 2010-12-02 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Involvement of the distal Arg residue in Cl- binding of midge larval haemoglobin
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3FJZ
 
 | | E. coli EPSP synthase (T97I) liganded with S3P and glyphosate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, N-(phosphonomethyl)glycine, ... | | Authors: | Schonbrunn, E. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Glyphosate Resistance Resulting from the Double Mutation Thr97 -> Ile and Pro101 -> Ser in 5-Enolpyruvylshikimate-3-phosphate Synthase from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
2OSF
 
 | | Inhibition of Carbonic Anhydrase II by Thioxolone: A Mechanistic and Structural Study | | Descriptor: | 4-MERCAPTOBENZENE-1,3-DIOL, Carbonic anhydrase 2, S-(2,4-dihydroxyphenyl) hydrogen thiocarbonate, ... | | Authors: | Albert, A.B, Caroli, G, Govindasamy, L, Agbandje-McKenna, M, McKenna, R, Tripp, B.C. | | Deposit date: | 2007-02-05 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of carbonic anhydrase II by thioxolone: a mechanistic and structural study.
Biochemistry, 47, 2008
|
|
1YKC
 
 | |
3FK1
 
 | | E. coli EPSP synthase (TIPS mutation) liganded with S3P and glyphosate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, N-(phosphonomethyl)glycine, ... | | Authors: | Schonbrunn, E. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Glyphosate Resistance Resulting from the Double Mutation Thr97 -> Ile and Pro101 -> Ser in 5-Enolpyruvylshikimate-3-phosphate Synthase from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3ARL
 
 | | Cl- binding hemoglobin component V form Propsilocerus akamusi under 500 mM NaCl at pH 5.5 | | Descriptor: | CHLORIDE ION, Hemoglobin V, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kuwada, T, Hasegawa, T, Takagi, T, Shishikura, F. | | Deposit date: | 2010-12-02 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Involvement of the distal Arg residue in Cl- binding of midge larval haemoglobin
Acta Crystallogr.,Sect.D, 67, 2011
|
|
7EOQ
 
 | |
7EOT
 
 | |
7EOU
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glycine/glutamate/GNE-6901/9-AA bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, 9-AMINOACRIDINE, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2021-04-22 | | Release date: | 2021-06-30 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Gating mechanism and a modulatory niche of human GluN1-GluN2A NMDA receptors.
Neuron, 109, 2021
|
|
