7PW0
 
 | |
6HNI
 
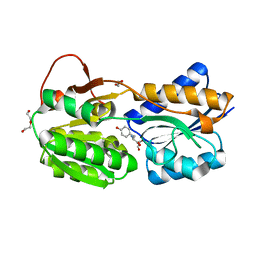 | | The ligand-bound, closed structure of CD0873, a substrate binding protein with adhesive properties from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, ABC-type transport system, sugar-family extracellular solute-binding protein, ... | | Authors: | Bradshaw, W.J, Kovacs-Simon, A, Harmer, N.J, Michell, S.L, Acharya, K.R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Molecular features of lipoprotein CD0873: A potential vaccine against the human pathogenClostridioides difficile.
J.Biol.Chem., 294, 2019
|
|
7KPF
 
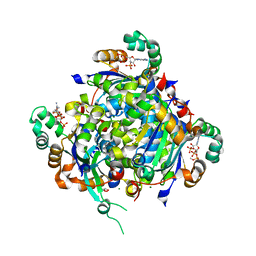 | | NME2 bound to myristoyl-CoA | | Descriptor: | GLYCEROL, MAGNESIUM ION, Nucleoside diphosphate kinase B, ... | | Authors: | Price, I.R, Lin, H. | | Deposit date: | 2020-11-11 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A Chemical Proteomic Approach reveals the regulation of NME1/2 and cancer metastasis by Long-Chain Fatty Acyl Coenzyme A
To Be Published
|
|
7PVT
 
 | |
7Z94
 
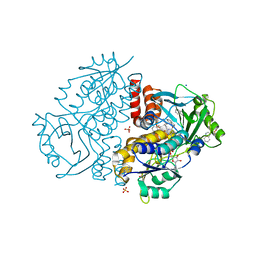 | | Crystal structure of Variovorax paradoxus indole monooxygenase (VpIndA1) in complex with indole | | Descriptor: | DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, INDOLE, ... | | Authors: | Kratky, J, Weisse, R, Strater, N. | | Deposit date: | 2022-03-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Mechanistic Studies on Substrate and Stereoselectivity of the Indole Monooxygenase VpIndA1: New Avenues for Biocatalytic Epoxidations and Sulfoxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8RV4
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 2 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-2-phenyl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
6ZRV
 
 | | Crystal Structure of Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-W175F) in Complex with an Inhibitory Nanobody (VHH-s-a93, Nb93) | | Descriptor: | Plasminogen activator inhibitor 1, VHH-s-a93 (Nb93) | | Authors: | Sillen, M, Weeks, S.D, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2020-07-15 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Insights into the Mechanism of a Nanobody That Stabilizes PAI-1 and Modulates Its Activity.
Int J Mol Sci, 21, 2020
|
|
7YYF
 
 | |
7KOB
 
 | | Crystal structure of antigen 43b from Escherichia coli CFT073 | | Descriptor: | Antigen 43b, GLYCEROL | | Authors: | Martinez-Ortiz, G.C, Hor, L, Paxman, J.J, Heras, B. | | Deposit date: | 2020-11-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Variation of Antigen 43 self-association modulates bacterial compacting within aggregates and biofilms.
NPJ Biofilms Microbiomes, 8, 2022
|
|
6ZS3
 
 | | Crystal structure of the fifth bromodomain of human protein polybromo-1 in complex with 2-(6-amino-5-(piperazin-1-yl)pyridazin-3-yl)phenol | | Descriptor: | 1,2-ETHANEDIOL, 2-(6-azanyl-5-piperazin-4-ium-1-yl-pyridazin-3-yl)phenol, Protein polybromo-1 | | Authors: | Preuss, F, Joerger, A.C, Wanior, M, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
6BCT
 
 | | I-LtrI E184D bound to non-cognate C4 substrate (pre-cleavage complex) | | Descriptor: | CALCIUM ION, DNA (26-MER), DNA (27-MER), ... | | Authors: | Brown, C, Zhang, K, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases.
Nucleic Acids Res., 46, 2018
|
|
7KOH
 
 | | Crystal structure of antigen 43 from Escherichia coli EDL933 | | Descriptor: | Antigen 43, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vo, J.L, Paxman, J.J, Heras, B. | | Deposit date: | 2020-11-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Variation of Antigen 43 self-association modulates bacterial compacting within aggregates and biofilms.
NPJ Biofilms Microbiomes, 8, 2022
|
|
7PVW
 
 | |
7KO9
 
 | | Crystal structure of antigen 43 from uropathogenic Escherichia coli UTI89 | | Descriptor: | AidA-I family adhesin, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vo, J.L, Hor, L, Paxman, J.J, Heras, B. | | Deposit date: | 2020-11-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Variation of Antigen 43 self-association modulates bacterial compacting within aggregates and biofilms.
NPJ Biofilms Microbiomes, 8, 2022
|
|
7PQI
 
 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PVY
 
 | |
8RV9
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 6 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-2-chloranyl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
6HQ9
 
 | | Crystal structure of the Tudor domain of human ERCC6-L2 | | Descriptor: | DNA excision repair protein ERCC-6-like 2 | | Authors: | Newman, J.A, Gavard, A.E, Nathan, W.J, Pinkas, D.M, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-09-24 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Crystal structure of the Tudor domain of human ERCC6-L2
To Be Published
|
|
6HQD
 
 | | Cytochrome P450-153 from Pseudomonas sp. 19-rlim | | Descriptor: | Cytochrome P450, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fiorentini, F, Mattevi, A. | | Deposit date: | 2018-09-24 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Extreme Structural Plasticity in the CYP153 Subfamily of P450s Directs Development of Designer Hydroxylases.
Biochemistry, 57, 2018
|
|
7PQM
 
 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with EBL2888 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PQL
 
 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with EBL2704 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1R)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
6HQW
 
 | |
6HMQ
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE BENZOTRIAZOLE-TYPE INHIBITOR MB002 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(4,5,6,7-tetrabromo-1H-benzotriazol-1-yl)propan-1-ol, ... | | Authors: | Niefind, K, Lindenblatt, D, Applegate, V.M, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
7Z4X
 
 | |
8RZC
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 11 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-imidazol-1-yl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
